The Omics Complexity in Sepsis: The Limits of the Personalized Medicine Approach
- PMID: 38540968
- PMCID: PMC10971110
- DOI: 10.3390/jpm14030225
The Omics Complexity in Sepsis: The Limits of the Personalized Medicine Approach
Abstract
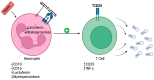
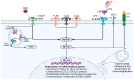
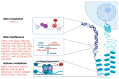
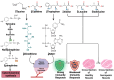
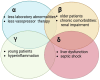
Sepsis is one of the most common causes of morbidity and mortality worldwide. Despite the remarkable advances in modern medicine throughout the last century, the mortality rates associated with sepsis have remained significantly elevated, both in high- and low-income countries. The main difficulty in the diagnosis and treatment of septic patients is the tremendous heterogeneity of this condition. The vast heterogeneity that characterizes sepsis ranges from the clinical presentation to the biological aspects of the disease. Evidence-based medicine approaches sepsis as a homogenous syndrome and does not consider the individual discrepancies between septic patients. This approach may contribute to the poor outcomes of septic patients. In recent years, personalized medicine has gained significant interest. This novel form of medicine underlines the importance of understanding the genetic, epigenetic, and molecular basis of a disease in order to provide a more tailored approach for the patient. The study of "omics", such as cytomics, genomics, epigenomics, transcriptomics, proteomics, and metabolomics, provides a deeper comprehension of the complex interactions between the host, the disease, and the environment. The aim of this review is to summarize the potential role of a personalized approach in sepsis management, considering the interactions between various "omics".
Keywords: clinical phenotypes; cytomics; epigenomics; genomics; metabolomics; proteomics; sepsis; septic shock; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Singer M., Deutschman C.S., Seymour C., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) J. Am. Med. Assoc. 2016;315:801–810. doi: 10.1001/jama.2016.0287. - DOI - PMC - PubMed
-
- Fleischmann C., Scherag A., Adhikari N.K.J., Hartog C.S., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K. Assessment of Global Incidence and Mortality of Hospital-Treated Sepsis Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. - DOI - PubMed
-
- Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

