The DUX4-HIF1α Axis in Murine and Human Muscle Cells: A Link More Complex Than Expected
- PMID: 38542301
- PMCID: PMC10969790
- DOI: 10.3390/ijms25063327
The DUX4-HIF1α Axis in Murine and Human Muscle Cells: A Link More Complex Than Expected
Abstract
FacioScapuloHumeral muscular Dystrophy (FSHD) is one of the most prevalent inherited muscle disorders and is linked to the inappropriate expression of the DUX4 transcription factor in skeletal muscles. The deregulated molecular network causing FSHD muscle dysfunction and pathology is not well understood. It has been shown that the hypoxia response factor HIF1α is critically disturbed in FSHD and has a major role in DUX4-induced cell death. In this study, we further explored the relationship between DUX4 and HIF1α. We found that the DUX4 and HIF1α link differed according to the stage of myogenic differentiation and was conserved between human and mouse muscle. Furthermore, we found that HIF1α knockdown in a mouse model of DUX4 local expression exacerbated DUX4-mediated muscle fibrosis. Our data indicate that the suggested role of HIF1α in DUX4 toxicity is complex and that targeting HIF1α might be challenging in the context of FSHD therapeutic approaches.
Keywords: DUX4; FSHD; HIF1α; myogenesis; skeletal muscle.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
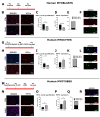
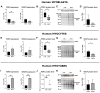
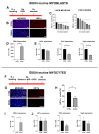
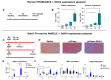

References
-
- Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O., et al. DUX4, a Candidate Gene of Facioscapulohumeral Muscular Dystrophy, Encodes a Transcriptional Activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. - DOI - PMC - PubMed
-
- Lemmers R.J.L.F., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. - DOI - PMC - PubMed
-
- Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. DUX4 Activates Germline Genes, Retroelements and Immune-Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- FC 29703/FRIA doctoral fellowship from the National Fund for Scientific Research (F.R.S - FNRS), Belgium
- 21GRO-PG12-0530/Muscular Dystrophy UK
- J 4435/FWF_/Austrian Science Fund FWF/Austria
- Amis FSH/French non-profit organization AMIS FSH (France)
- ABMM/Association Belge contre les Maladies neuro-Musculaires (ABMM, Belgium)
LinkOut - more resources
Full Text Sources

