Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Panax ginseng: Evidence for the Role of PgIAA02 in Lateral Root Development
- PMID: 38542445
- PMCID: PMC10971203
- DOI: 10.3390/ijms25063470
Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Panax ginseng: Evidence for the Role of PgIAA02 in Lateral Root Development
Abstract
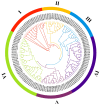
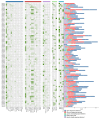
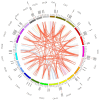
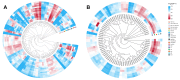
Panax ginseng C. A. Meyer (Ginseng) is one of the most used traditional Chinese herbal medicines, with its roots being used as the main common medicinal parts; its therapeutic potential has garnered significant attention. AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) is a family of early auxin-responsive genes capable of regulating root development in plants through the auxin signaling pathway. In the present study, 84 Aux/IAA genes were identified from the ginseng genome and their complexity and diversity were determined through their protein domains, phylogenetic relationships, gene structures, and cis-acting element predictions. Phylogenetic analyses classified PgIAA into six subgroups, with members in the same group showing greater sequence similarity. Analyses of interspecific collinearity suggest that segmental duplications likely drove the evolution of PgIAA genes, followed by purifying selection. An analysis of cis-regulatory elements suggested that PgIAA family genes may be involved in the regulation of plant hormones. RNA-seq data show that the expression pattern of Aux/IAA genes in Ginseng is tissue-specific, and PgIAA02 and PgIAA36 are specifically highly expressed in lateral, fibrous, and arm roots, suggesting their potential function in root development. The PgIAA02 overexpression lines exhibited an inhibition of lateral root growth in Ginseng. In addition, yeast two-hybrid and subcellular localization experiments showed that PgIAA02 interacted with PgARF22/PgARF36 (ARF: auxin response factor) in the nucleus and participated in the biological process of root development. The above results lay the foundation for an in-depth study of Aux/IAA and provide preliminary information for further research on the role of the Aux/IAA gene family in the root development of Ginseng.
Keywords: Aux/IAA gene family; Panax ginseng; auxin-responsive genes; evolution; expression pattern analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Genome-wide identification and expression pattern analysis of the Aux/IAA (auxin/indole-3-acetic acid) gene family in alfalfa (Medicago sativa) and the potential functions under drought stress.BMC Genomics. 2024 Apr 18;25(1):382. doi: 10.1186/s12864-024-10313-2. BMC Genomics. 2024. PMID: 38637768 Free PMC article.
-
Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa).Funct Integr Genomics. 2006 Jan;6(1):47-59. doi: 10.1007/s10142-005-0005-0. Epub 2005 Oct 1. Funct Integr Genomics. 2006. PMID: 16200395
-
Genome-Wide Identification, Expression and Interaction Analysis of ARF and AUX/IAA Gene Family in Soybean.Front Biosci (Landmark Ed). 2022 Aug 19;27(8):251. doi: 10.31083/j.fbl2708251. Front Biosci (Landmark Ed). 2022. PMID: 36042185
-
Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function.Int J Mol Sci. 2018 Jan 16;19(1):259. doi: 10.3390/ijms19010259. Int J Mol Sci. 2018. PMID: 29337875 Free PMC article. Review.
-
Advances in structure and function of auxin response factor in plants.J Integr Plant Biol. 2023 Mar;65(3):617-632. doi: 10.1111/jipb.13392. Epub 2022 Dec 31. J Integr Plant Biol. 2023. PMID: 36263892 Review.
Cited by
-
Genome-Wide Analysis of the Auxin/Indoleacetic Acid (Aux/IAA) Gene Family in Autopolyploid Sugarcane (Saccharum spontaneum).Int J Mol Sci. 2024 Jul 8;25(13):7473. doi: 10.3390/ijms25137473. Int J Mol Sci. 2024. PMID: 39000581 Free PMC article.
-
Genome-wide identification of the GRAS gene family and evidence for the involvement of PgGRAS48 in main root development in Panax ginseng.Front Plant Sci. 2025 Jun 13;16:1603268. doi: 10.3389/fpls.2025.1603268. eCollection 2025. Front Plant Sci. 2025. PMID: 40584861 Free PMC article.
-
Genome-Wide Identification and Characterization of the Aux/IAA Gene Family in Strawberry Species.Plants (Basel). 2024 Oct 21;13(20):2940. doi: 10.3390/plants13202940. Plants (Basel). 2024. PMID: 39458886 Free PMC article.
References
-
- Thimann K.V., Went F.W. On the chemical nature of the root forming hormone. Proc. K. Ned. Akad. Van Wet. 1934;37:456–459.
-
- Yu C.L., Sun C.D., Shen C.J., Wang S.K., Liu F., Liu Y., Chen Y.L., Li C.Y., Qian Q., Aryal B., et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryzasativa L.) Plant J. 2015;83:818–830. doi: 10.1111/tpj.12929. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

