Age-Dependent Inflammatory Microenvironment Mediates Alveolar Regeneration
- PMID: 38542450
- PMCID: PMC10970842
- DOI: 10.3390/ijms25063476
Age-Dependent Inflammatory Microenvironment Mediates Alveolar Regeneration
Abstract
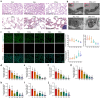
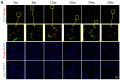
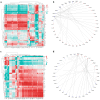
Lung aging triggers the onset of various chronic lung diseases, with alveolar repair being a key focus for alleviating pulmonary conditions. The regeneration of epithelial structures, particularly the differentiation from type II alveolar epithelial (AT2) cells to type I alveolar epithelial (AT1) cells, serves as a prominent indicator of alveolar repair. Nonetheless, the precise role of aging in impeding alveolar regeneration and its underlying mechanism remain to be fully elucidated. Our study employed histological methods to examine lung aging effects on structural integrity and pathology. Lung aging led to alveolar collapse, disrupted epithelial structures, and inflammation. Additionally, a relative quantification analysis revealed age-related decline in AT1 and AT2 cells, along with reduced proliferation and differentiation capacities of AT2 cells. To elucidate the mechanisms underlying AT2 cell functional decline, we employed transcriptomic techniques and revealed a correlation between inflammatory factors and genes regulating proliferation and differentiation. Furthermore, a D-galactose-induced senescence model in A549 cells corroborated our omics experiments and confirmed inflammation-induced cell cycle arrest and a >30% reduction in proliferation/differentiation. Physiological aging-induced chronic inflammation impairs AT2 cell functions, hindering tissue repair and promoting lung disease progression. This study offers novel insights into chronic inflammation's impact on stem cell-mediated alveolar regeneration.
Keywords: AT2; alveolar regeneration; differentiation; inflammation; proliferation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- American Lung Association Trends in Pneumonia and Influenza Morbidity and Mortality. [(accessed on 10 July 2023)]. Available online: https://www.lung.org/getmedia/98f088b5-3fd7-4c43-a490-ba8f4747bd4d/pi-tr....
-
- Angelidis I., Simon L.M., Fernandez I.E., Strunz M., Mayr C.H., Greiffo F.R., Tsitsiridis G., Ansari M., Graf E., Strom T.M., et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

