Multi-Omics Analysis of Gut Microbiota and Host Transcriptomics Reveal Dysregulated Immune Response and Metabolism in Young Adults with Irritable Bowel Syndrome
- PMID: 38542485
- PMCID: PMC10970623
- DOI: 10.3390/ijms25063514
Multi-Omics Analysis of Gut Microbiota and Host Transcriptomics Reveal Dysregulated Immune Response and Metabolism in Young Adults with Irritable Bowel Syndrome
Abstract
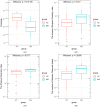
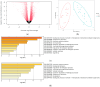
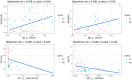
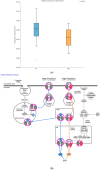
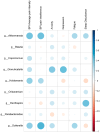
The integrated dysbiosis of gut microbiota and altered host transcriptomics in irritable bowel syndrome (IBS) is yet to be known. This study investigated the associations among gut microbiota and host transcriptomics in young adults with IBS. Stool and peripheral blood samples from 20 IBS subjects and 21 healthy controls (HCs) collected at the baseline visit of an RCT were sequenced to depict the gut microbiota and transcriptomic profiles, respectively. The diversities, composition, and predicted metabolic pathways of gut microbiota significantly differed between IBS subjects and HCs. Nine genera were significantly abundant in IBS stool samples, including Akkermansia, Blautia, Coprococcus, Granulicatella, Holdemania, Oribacterium, Oscillospira, Parabacteroides, and Sutterella. There were 2264 DEGs found between IBS subjects and HCs; 768 were upregulated, and 1496 were downregulated in IBS participants compared with HCs. The enriched gene ontology included the immune system process and immune response. The pathway of antigen processing and presentation (hsa04612) in gut microbiota was also significantly different in the RNA-seq data. Akkermansia, Blautia, Holdemania, and Sutterella were significantly correlated with ANXA2P2 (upregulated, positive correlations), PCSK1N (downregulated, negative correlations), and GLTPD2 (downregulated, negative correlations). This study identified the dysregulated immune response and metabolism in IBS participants revealed by the altered gut microbiota and transcriptomic profiles.
Keywords: gene expression profiling; gut microbiome; immunity; irritable bowel syndrome; metabolic processes; multi-omics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Multiomics Analysis Reveals Gut Virome-Bacteria-Metabolite Interactions and Their Associations with Symptoms in Patients with IBS-D.Viruses. 2024 Jun 29;16(7):1054. doi: 10.3390/v16071054. Viruses. 2024. PMID: 39066219 Free PMC article.
-
Integrated fecal microbiome-metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome.Gut Microbes. 2022 Jan-Dec;14(1):2063016. doi: 10.1080/19490976.2022.2063016. Gut Microbes. 2022. PMID: 35446234 Free PMC article.
-
Altered metabolism of bile acids correlates with clinical parameters and the gut microbiota in patients with diarrhea-predominant irritable bowel syndrome.World J Gastroenterol. 2020 Dec 7;26(45):7153-7172. doi: 10.3748/wjg.v26.i45.7153. World J Gastroenterol. 2020. PMID: 33362374 Free PMC article.
-
The gut microbiome and irritable bowel syndrome: State of art review.Arab J Gastroenterol. 2018 Sep;19(3):136-141. doi: 10.1016/j.ajg.2018.02.008. Epub 2018 Jun 20. Arab J Gastroenterol. 2018. PMID: 29935865 Review.
-
Gut microbiota and irritable bowel syndrome.J Dig Dis. 2023 May;24(5):312-320. doi: 10.1111/1751-2980.13204. Epub 2023 Aug 4. J Dig Dis. 2023. PMID: 37458142 Review.
Cited by
-
Efficacy of Probiotics in the Management of Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis.Cureus. 2024 Dec 18;16(12):e75954. doi: 10.7759/cureus.75954. eCollection 2024 Dec. Cureus. 2024. PMID: 39830577 Free PMC article. Review.
-
Antioxidant Peptides Derived from Woody Oil Resources: Mechanisms of Redox Protection and Emerging Therapeutic Opportunities.Pharmaceuticals (Basel). 2025 Jun 4;18(6):842. doi: 10.3390/ph18060842. Pharmaceuticals (Basel). 2025. PMID: 40573238 Free PMC article. Review.
-
Gut microbiota influences colorectal cancer through immune cell interactions: a Mendelian randomization study.Discov Oncol. 2025 May 13;16(1):747. doi: 10.1007/s12672-025-02486-3. Discov Oncol. 2025. PMID: 40358736 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

