The Effects of Selected Extraction Methods and Natural Deep Eutectic Solvents on the Recovery of Active Principles from Aralia elata var. mandshurica (Rupr. & Maxim.) J. Wen: A Non-Targeted Metabolomics Approach
- PMID: 38543141
- PMCID: PMC10974460
- DOI: 10.3390/ph17030355
The Effects of Selected Extraction Methods and Natural Deep Eutectic Solvents on the Recovery of Active Principles from Aralia elata var. mandshurica (Rupr. & Maxim.) J. Wen: A Non-Targeted Metabolomics Approach
Abstract
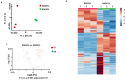
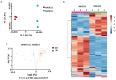
The methods and solvents employed in routine extraction protocols essentially impact the composition of the resulting extracts, i.e., the relative abundances of individual biologically active metabolites and the quality and stability of the isolates. Natural deep eutectic solvents (NADESs) represent a new class of environmentally friendly solvents, which are recognized as promising extractants alternative to conventional organic liquids. However, their relative efficiencies when applied in different extraction workflows are still poorly characterized. Therefore, here, we compare the potential of three extraction methods for the extraction of biologically active natural products from Aralia elata var. mandshurica with selected natural deep eutectic solvents (NADESs) using a non-targeted metabolomics approach. The non-targeted metabolite profiling relied on reversed-phase ultra-high-performance liquid chromatography-high-resolution mass spectrometry (RP-UHPLC-HR-MS). The roots of A. elata were extracted by maceration, ultrasound-assisted extraction (UAE), and vibrocavitation-assisted extraction (VAE). Principal component analysis (PCA) revealed a clear separation of the extracts obtained with the three extraction methods employed with NADES1 (choline chloride/malic acid) and NADES2 (sorbitol/malic acid/water). Based on the results of the hierarchical clustering analysis obtained for the normalized relative abundances of individual metabolites and further statistical evaluation with the t-test, it could be concluded that NADES1 showed superior extraction efficiency for all the protocols addressed. Therefore, this NADES was selected to compare the efficiencies of the three extraction methods in more detail. PCA followed by the t-test yielded only 3 metabolites that were more efficiently extracted by maceration, whereas 46 compounds were more abundant in the extracts obtained by VAE. When VAE and UAE were compared, 108 metabolites appeared to be more abundant in the extracts obtained by VAE, whereas only 1 metabolite was more efficiently recovered by UAE. These facts clearly indicate the advantage of the VAE method over maceration and UAE. Seven of the twenty-seven metabolites tentatively identified by tandem mass spectrometry (MS/MS) were found in the roots of A. elata for the first time. Additional studies are necessary to understand the applicability of VAE for the extraction of other plant materials.
Keywords: Aralia elata; NADES; extraction; natural deep eutectic solvents; non-targeted metabolics profiling; ultrasound-assisted extraction; vibrocavitation-assisted extraction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Fuad F.M., Nadzir M.M., Harun A. Hydrophilic natural deep eutectic solvent: A review on physicochemical properties and extractability of bioactive compounds. J. Mol. Liq. 2021;339:116923. doi: 10.1016/j.molliq.2021.116923. - DOI
-
- Da Silva R.F., Carneiro C.N., Cheila C.B., de Sousa C.B.D.C., Gomez F.J.V., Espino M., Boiteux J., Fernández M.D.L.Á., Silva M.F., Dias F.D.S. Sustainable extraction bioactive compounds procedures in medicinal plants based on the principles of green analytical chemistry: A review. Microchem. J. 2022;175:107184. doi: 10.1016/j.microc.2022.107184. - DOI
LinkOut - more resources
Full Text Sources

