Genome-Based Taxonomy of Species in the Pseudomonas syringae and Pseudomonas lutea Phylogenetic Groups and Proposal of Pseudomonas maioricensis sp. nov., Isolated from Agricultural Soil
- PMID: 38543511
- PMCID: PMC10974278
- DOI: 10.3390/microorganisms12030460
Genome-Based Taxonomy of Species in the Pseudomonas syringae and Pseudomonas lutea Phylogenetic Groups and Proposal of Pseudomonas maioricensis sp. nov., Isolated from Agricultural Soil
Abstract
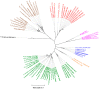
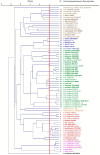
Species in the phylogenetic group Pseudomonas syringae are considered the most relevant plant pathogenic bacteria, but their taxonomy is still controversial. Twenty named species are validated in the current taxonomy of this group and in recent years many strains have been genome-sequenced, putative new species have been proposed and an update in the taxonomy is needed. A taxonomic study based on the core-genome phylogeny, genomic indices (ANI and GGDC) and gene content (phyletic pattern and Jaccard index) have been applied to clarify the taxonomy of the group. A phylogenomic analysis demonstrates that at least 50 phylogenomic species can be delineated within the group and that many strains whose genomes have been deposited in the databases are not correctly classified at the species level. Other species names, like "Pseudomonas coronafaciens", have been proposed but are not validated yet. One of the putative new species is taxonomically described, and the name Pseudomonas maioricensis sp. nov. is proposed. The taxonomies of Pseudomonas avellanae and Pseudomonas viridiflava are discussed in detail as case studies. Correct strain identification is a prerequisite for many studies, and therefore, criteria are given to facilitate identification.
Keywords: Pseudomonas maioricensis; Pseudomonas syringae; phylogenomics; taxonomy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Bashan Y., Okon Y., Henis Y. Infection studies of Pseudomonas tomato, causal agent of bacterial speck of tomato. Phytoparasitica. 1978;6:135–143. doi: 10.1007/BF02981213. - DOI
-
- Stefani E., Caffier D., Fiore N. The economic impact of the bacterial blight of soybean under European agroclimatic conditions. J. Plant Pathol. 1998;80:211–221.
-
- Fatmi M.B., Collmer A., Sante Iacobellis N., Mansfield J.W., Murillo J., Schaad N.W., Ullrich M. Pseudomonas syringae Pathovars and Related Pathogens—Identification, Epidemiology and Genomics. Springer; Dordrecht, The Netherlands: 2008. pp. 1–433.
Grants and funding
LinkOut - more resources
Full Text Sources

