A Pseudovirus-Based Neutralization Assay for SARS-CoV-2 Variants: A Rapid, Cost-Effective, BSL-2-Based High-Throughput Assay Useful for Vaccine Immunogenicity Evaluation
- PMID: 38543552
- PMCID: PMC10972517
- DOI: 10.3390/microorganisms12030501
A Pseudovirus-Based Neutralization Assay for SARS-CoV-2 Variants: A Rapid, Cost-Effective, BSL-2-Based High-Throughput Assay Useful for Vaccine Immunogenicity Evaluation
Abstract
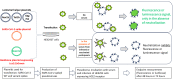
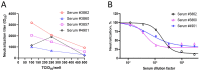
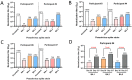
Neutralizing antibody responses from COVID-19 vaccines are pivotal in conferring protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Effective COVID-19 vaccines and assays measuring neutralizing antibodies against emerging variants (i.e., XBB.1.5, XBB.1.16, and XBB.2.3) are needed. The use of biosafety level (BSL)-3 laboratories for live virus assays results in higher costs and a longer turnaround time; therefore, a BSL-2-based pseudovirus neutralization assay (PNT) was developed. The pseudoviruses were produced by cotransfecting cells with plasmids encoding a lentiviral backbone-expressing luciferase reporter; non-surface proteins for lentiviral production; and ancestral or Omicron (BA.1 and BA.5) SARS-CoV-2 spike (S) proteins. The PNT was developed and optimized in dose and kinetics experiments. The representative serum samples (COVID-19-convalescent or NVX-CoV2373-vaccinated participants enrolled in the 2019nCoV-101 trial) demonstrated a wide dynamic range. The neutralization data showed robust correlation with validated anti-recombinant spike IgG levels and angiotensin-converting enzyme 2 inhibition titers (ancestral). This assay is suitable for measurement of the neutralization ability in clinical samples from individuals infected with SARS-CoV-2 or immunized with a COVID-19 vaccine. The results suggest that this PNT provides a lower cost, high-throughput, rapid turnaround alternative to BSL-3-based microneutralization assays and enables the discovery and development of effective vaccines against emerging variants.
Keywords: COVID-19; SARS-CoV-2; XBB.1.16; XBB.1.5; XBB.2.3; assay validation; correlate of protection; immunogenicity; neutralizing antibody titers; pseudovirus-based neutralization assays.
Conflict of interest statement
All the authors are employees and stockholders of Novavax, Inc. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Besides, the authors declare that this study received funding from Novavax, Inc. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Figures







References
-
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. - DOI - PubMed
-
- World Health Organization Tracking SARS-CoV-2 variants. [(accessed on 31 January 2024)]. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants.
-
- Stanford University Coronavirus Antiviral and Resistance Database SARS-CoV-2 Variants. [(accessed on 31 January 2024)]. Available online: https://covdb.stanford.edu/variants/omicron_xbb/
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

