A Mini-Review on the Common Antiviral Drug Targets of Coronavirus
- PMID: 38543651
- PMCID: PMC10974577
- DOI: 10.3390/microorganisms12030600
A Mini-Review on the Common Antiviral Drug Targets of Coronavirus
Abstract
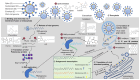
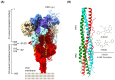
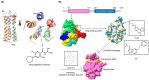
Coronaviruses in general are a zoonotic pathogen with significant cross-species transmission. They are widely distributed in nature and have recently become a major threat to global public health. Vaccines are the preferred strategy for the prevention of coronaviruses. However, the rapid rate of virus mutation, large number of prevalent strains, and lag in vaccine development contribute to the continuing frequent occurrence of coronavirus diseases. There is an urgent need for new antiviral strategies to address coronavirus infections effectively. Antiviral drugs are important in the prevention and control of viral diseases. Members of the genus coronavirus are highly similar in life-cycle processes such as viral invasion and replication. These, together with the high degree of similarity in the protein sequences and structures of viruses in the same genus, provide common targets for antiviral drug screening of coronaviruses and have led to important advances in recent years. In this review, we summarize the pathogenic mechanisms of coronavirus, common drugs targeting coronavirus entry into host cells, and common drug targets against coronaviruses based on biosynthesis and on viral assembly and release. We also describe the common targets of antiviral drugs against coronaviruses and the progress of antiviral drug research. Our aim is to provide a theoretical basis for the development of antiviral drugs and to accelerate the development and utilization of commonly used antiviral drugs in China.
Keywords: broad-spectrum; common antivirals; common mechanism; coronavirus; virus targets.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
Cell-based antiviral screening against coronaviruses: developing virus-specific and broad-spectrum inhibitors.Antiviral Res. 2014 Jan;101:105-12. doi: 10.1016/j.antiviral.2013.11.004. Epub 2013 Nov 20. Antiviral Res. 2014. PMID: 24269477 Free PMC article. Review.
-
Biologically active compounds from marine organisms in the strategies for combating coronaviruses.AIMS Microbiol. 2020 Dec 7;6(4):470-494. doi: 10.3934/microbiol.2020028. eCollection 2020. AIMS Microbiol. 2020. PMID: 33364539 Free PMC article. Review.
-
Adaptive Mutation in the Main Protease Cleavage Site of Feline Coronavirus Renders the Virus More Resistant to Main Protease Inhibitors.J Virol. 2022 Sep 14;96(17):e0090722. doi: 10.1128/jvi.00907-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000844 Free PMC article.
Cited by
-
Rotavirus Reverse Genetics Systems and Oral Vaccine Delivery Vectors for Mucosal Vaccination.Microorganisms. 2025 Jul 4;13(7):1579. doi: 10.3390/microorganisms13071579. Microorganisms. 2025. PMID: 40732089 Free PMC article. Review.
-
Arylamines QSAR-Based Design and Molecular Dynamics of New Phenylthiophene and Benzimidazole Derivatives with Affinity for the C111, Y268, and H73 Sites of SARS-CoV-2 PLpro Enzyme.Pharmaceuticals (Basel). 2024 May 9;17(5):606. doi: 10.3390/ph17050606. Pharmaceuticals (Basel). 2024. PMID: 38794177 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

