Discordant Antigenic Properties of Soluble and Virion SARS-CoV-2 Spike Proteins
- PMID: 38543772
- PMCID: PMC10974403
- DOI: 10.3390/v16030407
Discordant Antigenic Properties of Soluble and Virion SARS-CoV-2 Spike Proteins
Abstract
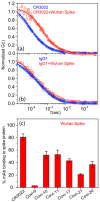
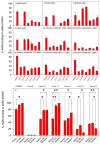
Efforts to develop vaccine and immunotherapeutic countermeasures against the COVID-19 pandemic focus on targeting the trimeric spike (S) proteins of SARS-CoV-2. Vaccines and therapeutic design strategies must impart the characteristics of virion S from historical and emerging variants onto practical constructs such as soluble, stabilized trimers. The virus spike is a heterotrimer of two subunits: S1, which includes the receptor binding domain (RBD) that binds the cell surface receptor ACE2, and S2, which mediates membrane fusion. Previous studies suggest that the antigenic, structural, and functional characteristics of virion S may differ from current soluble surrogates. For example, it was reported that certain anti-glycan, HIV-1 neutralizing monoclonal antibodies bind soluble SARS-CoV-2 S but do not neutralize SARS-CoV-2 virions. In this study, we used single-molecule fluorescence correlation spectroscopy (FCS) under physiologically relevant conditions to examine the reactivity of broadly neutralizing and non-neutralizing anti-S human monoclonal antibodies (mAbs) isolated in 2020. Binding efficiency was assessed by FCS with soluble S trimers, pseudoviruses and inactivated wild-type virions representing variants emerging from 2020 to date. Anti-glycan mAbs were tested and compared. We find that both anti-S specific and anti-glycan mAbs exhibit variable but efficient binding to a range of stabilized, soluble trimers. Across mAbs, the efficiencies of soluble S binding were positively correlated with reactivity against inactivated virions but not pseudoviruses. Binding efficiencies with pseudoviruses were generally lower than with soluble S or inactivated virions. Among neutralizing mAbs, potency did not correlate with binding efficiencies on any target. No neutralizing activity was detected with anti-glycan antibodies. Notably, the virion S released from membranes by detergent treatment gained more efficient reactivity with anti-glycan, HIV-neutralizing antibodies but lost reactivity with all anti-S mAbs. Collectively, the FCS binding data suggest that virion surfaces present appreciable amounts of both functional and nonfunctional trimers, with neutralizing anti-S favoring the former structures and non-neutralizing anti-glycan mAbs binding the latter. S released from solubilized virions represents a nonfunctional structure bound by anti-glycan mAbs, while engineered soluble trimers present a composite structure that is broadly reactive with both mAb types. The detection of disparate antigenicity and immunoreactivity profiles in engineered and virion-associated S highlight the value of single-virus analyses in designing future antiviral strategies against SARS-CoV-2.
Keywords: Covi-mAbs; SARS-CoV-2 spike; anti-HIV glycan mAbs; antigenicity; fluorescence correlation spectroscopy; in vitro binding assay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. - DOI - PMC - PubMed
-
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

