Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments
- PMID: 38543786
- PMCID: PMC10974462
- DOI: 10.3390/v16030421
Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments
Abstract
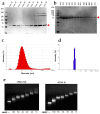
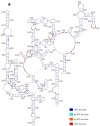
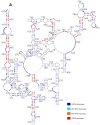
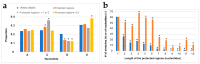
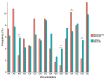
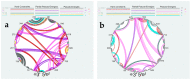
Influenza A viruses (IAVs) possess a segmented genome consisting of eight viral RNAs (vRNAs) associated with multiple copies of viral nucleoprotein (NP) and a viral polymerase complex. Despite the crucial role of RNA structure in IAV replication, the impact of NP binding on vRNA structure is not well understood. In this study, we employed SHAPE chemical probing to compare the structure of NS and M vRNAs of WSN IAV in various states: before the addition of NP, in complex with NP, and after the removal of NP. Comparison of the RNA structures before the addition of NP and after its removal reveals that NP, while introducing limited changes, remodels local structures in both vRNAs and long-range interactions in the NS vRNA, suggesting a potentially biologically relevant RNA chaperone activity. In contrast, NP significantly alters the structure of vRNAs in vRNA/NP complexes, though incorporating experimental data into RNA secondary structure prediction proved challenging. Finally, our results suggest that NP not only binds single-stranded RNA but also helices with interruptions, such as bulges or small internal loops, with a preference for G-poor and C/U-rich regions.
Keywords: NP; RNA chaperon; RNA structure; chemical probing; influenza A virus; nucleoprotein; vRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

