Recommendations for Uniform Variant Calling of SARS-CoV-2 Genome Sequence across Bioinformatic Workflows
- PMID: 38543795
- PMCID: PMC10975397
- DOI: 10.3390/v16030430
Recommendations for Uniform Variant Calling of SARS-CoV-2 Genome Sequence across Bioinformatic Workflows
Abstract
Genomic sequencing of clinical samples to identify emerging variants of SARS-CoV-2 has been a key public health tool for curbing the spread of the virus. As a result, an unprecedented number of SARS-CoV-2 genomes were sequenced during the COVID-19 pandemic, which allowed for rapid identification of genetic variants, enabling the timely design and testing of therapies and deployment of new vaccine formulations to combat the new variants. However, despite the technological advances of deep sequencing, the analysis of the raw sequence data generated globally is neither standardized nor consistent, leading to vastly disparate sequences that may impact identification of variants. Here, we show that for both Illumina and Oxford Nanopore sequencing platforms, downstream bioinformatic protocols used by industry, government, and academic groups resulted in different virus sequences from same sample. These bioinformatic workflows produced consensus genomes with differences in single nucleotide polymorphisms, inclusion and exclusion of insertions, and/or deletions, despite using the same raw sequence as input datasets. Here, we compared and characterized such discrepancies and propose a specific suite of parameters and protocols that should be adopted across the field. Consistent results from bioinformatic workflows are fundamental to SARS-CoV-2 and future pathogen surveillance efforts, including pandemic preparation, to allow for a data-driven and timely public health response.
Keywords: SARS-CoV-2; variant calling.
Conflict of interest statement
R.M., J.L., and E.S.M. are employees of, and stockholders in, Gilead Sciences, Inc. J.d.I and L.A.P. are employees of Vir Biotechnology, Inc. and may hold shares in Vir Biotechnology, Inc. L.A.P. is a former employee and shareholder of Regeneron Pharmaceuticals and is a member of the Scientific Advisory Board of the AI-driven Structure-enabled Antiviral Platform (ASAP). P.E. is an employee of, and holds stock or stock options in, Eli Lilly and Company. Courtney Copeland and Tré LaRosa are employees of Deloitte Consulting LLP and have indicated they have no conflicts of interest relevant to this article to disclose. The remaining authors declare no conflicts of interest.
Figures
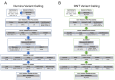
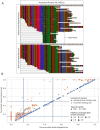




Update of
-
Towards increased accuracy and reproducibility in SARS-CoV-2 next generation sequence analysis for public health surveillance.bioRxiv [Preprint]. 2022 Nov 3:2022.11.03.515010. doi: 10.1101/2022.11.03.515010. bioRxiv. 2022. Update in: Viruses. 2024 Mar 11;16(3):430. doi: 10.3390/v16030430. PMID: 36380755 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

