Immune System Deficiencies Do Not Alter SARS-CoV-2 Evolutionary Rate but Favour the Emergence of Mutations by Extending Viral Persistence
- PMID: 38543811
- PMCID: PMC10974344
- DOI: 10.3390/v16030447
Immune System Deficiencies Do Not Alter SARS-CoV-2 Evolutionary Rate but Favour the Emergence of Mutations by Extending Viral Persistence
Abstract
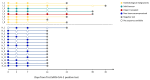
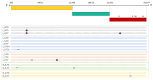
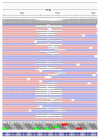
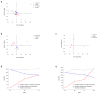
During the COVID-19 pandemic, immunosuppressed patients showed prolonged SARS-CoV-2 infections, with several studies reporting the accumulation of mutations in the viral genome. The weakened immune system present in these individuals, along with the effect of antiviral therapies, are thought to create a favourable environment for intra-host viral evolution and have been linked to the emergence of new viral variants which strongly challenged containment measures and some therapeutic treatments. To assess whether impaired immunity could lead to the increased instability of viral genomes, longitudinal nasopharyngeal swabs were collected from eight immunocompromised patients and fourteen non-immunocompromised subjects, all undergoing SARS-CoV-2 infection. Intra-host viral evolution was compared between the two groups through deep sequencing, exploiting a probe-based enrichment method to minimise the possibility of artefactual mutations commonly generated in amplicon-based methods, which heavily rely on PCR amplification. Although, as expected, immunocompromised patients experienced significantly longer infections, the acquisition of novel intra-host viral mutations was similar between the two groups. Moreover, a thorough analysis of viral quasispecies showed that the variability of viral populations in the two groups is comparable not only at the consensus level, but also when considering low-frequency mutations. This study suggests that a compromised immune system alone does not affect SARS-CoV-2 within-host genomic variability.
Keywords: SARS-CoV-2 genomic variability; immunocompromised subjects; intra-host; viral quasispecies.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. - DOI - PMC - PubMed
-
- Quaranta E.G., Fusaro A., Giussani E., D’Amico V., Varotto M., Pagliari M., Giordani M.T., Zoppelletto M., Merola F., Antico A., et al. SARS-CoV-2 Intra-Host Evolution during Prolonged Infection in an Immunocompromised Patient. Int. J. Infect. Dis. 2022;122:444–448. doi: 10.1016/j.ijid.2022.06.023. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

