High-Content Imaging-Based Assay for SARS-CoV-2-Neutralizing Antibodies
- PMID: 38543870
- PMCID: PMC10974448
- DOI: 10.3390/vaccines12030236
High-Content Imaging-Based Assay for SARS-CoV-2-Neutralizing Antibodies
Abstract
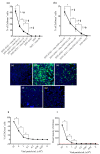
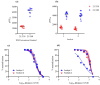
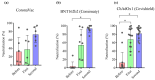
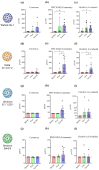
The COVID-19 pandemic and the consequent emergence of new SARS-CoV-2 variants of concern necessitates the determination of populational serum potency against the virus. Here, we standardized and validated an imaging-based method to quantify neutralizing antibodies against lentiviral particles expressing the spike glycoprotein (pseudovirus). This method was found to efficiently quantify viral titers based on ZsGreen-positive cells and detect changes in human serum neutralization capacity induced by vaccination with up to two doses of CoronaVac, Comirnaty, or Covishield vaccines. The imaging-based protocol was also used to quantify serum potency against pseudoviruses expressing spikes from Delta, Omicron BA.1.1.529, and BA.4/5. Our results revealed increases in serum potency after one and two doses of the vaccines evaluated and demonstrated that Delta and Omicron variants escape from antibody neutralization. The method presented herein represents a valuable tool for the screening of antibodies and small molecules capable of blocking viral entry and could be used to evaluate humoral immunity developed by different populations and for vaccine development.
Keywords: SARS-CoV-2; antibodies; high-content screening; neutralization; variants.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Vaccines—COVID-19 Vaccine Tracker. [(accessed on 26 December 2023)]. Available online: https://covid19.trackvaccines.org/vaccines/approved/#vaccine-list.
-
- Acharya C.B., Schrom J., Mitchell A.M., Coil D.A., Marquez C., Rojas S., Wang C.Y., Liu J., Pilarowski G., Solis L., et al. Viral Load among Vaccinated and Unvaccinated, Asymptomatic and Symptomatic Persons Infected with the SARS-CoV-2 Delta Variant. Open Forum Infect. Dis. 2022;9:ofac135. doi: 10.1093/ofid/ofac135. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

