Unveiling a Shield of Hope: A Novel Multiepitope-Based Immunogen for Cross-Serotype Cellular Defense against Dengue Virus
- PMID: 38543950
- PMCID: PMC10975250
- DOI: 10.3390/vaccines12030316
Unveiling a Shield of Hope: A Novel Multiepitope-Based Immunogen for Cross-Serotype Cellular Defense against Dengue Virus
Abstract
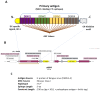
Dengue virus (DENV) infection continues to be a public health challenge, lacking a specific cure. Vaccination remains the primary strategy against dengue; however, existing live-attenuated vaccines display variable efficacy across four serotypes, influenced by host serostatus and age, and predominantly inducing humoral responses. To address this limitation, this study investigates a multiepitope-based immunogen designed to induce robust cellular immunity across all DENV serotypes. The chimeric immunogen integrates H-2d specific MHC-I binding T-cell epitopes derived from conserved domains within the DENV envelope protein. Immuno-informatics analyses supported its stability, non-allergenic nature, and strong MHC-I binding affinity as an antigen. To assess the immunogenicity of the multiepitope, it was expressed in murine bone-marrow-derived dendritic cells (BMDCs) that were used to prime mice. In this experimental model, simultaneous exposure to T-cell epitopes from all four DENV serotypes initiated distinct IFNγ-CD8 T-cell responses for different serotypes. These results supported the potential of the multiepitope construct as a vaccine candidate. While the optimization of the immunogen design remains a continuous pursuit, this proof-of-concept study provides a starting point for evaluating its protective efficacy against dengue infection in vivo. Moreover, our results support the development of a multiepitope vaccine that could trigger a pan-serotype anti-dengue CD8 response.
Keywords: MHC I-binding epitopes; cellular immunity; cross-serotype protection; dengue virus; immunization; memory T cells; multiepitope vaccine; vaccine candidate.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials

