Identifying of immune-associated genes for assessing the obesity-associated risk to the offspring in maternal obesity: A bioinformatics and machine learning
- PMID: 38544384
- PMCID: PMC10973700
- DOI: 10.1111/cns.14700
Identifying of immune-associated genes for assessing the obesity-associated risk to the offspring in maternal obesity: A bioinformatics and machine learning
Abstract
Background: Perinatal exposure to maternal obesity predisposes offspring to develop obesity later in life. Immune dysregulation in the hypothalamus, the brain center governing energy homeostasis, is pivotal in obesity development. This study aimed to identify key candidate genes associated with the risk of offspring obesity in maternal obesity.
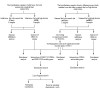
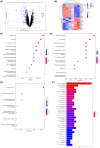
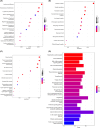
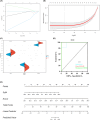
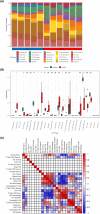
Methods: We obtained obesity-related datasets from the Gene Expression Omnibus (GEO) database. GSE135830 comprises gene expression data from the hypothalamus of mouse offspring in a maternal obesity model induced by a high-fat diet model (maternal high-fat diet (mHFD) group and maternal chow (mChow) group), while GSE127056 consists of hypothalamus microarray data from young adult mice with obesity (high-fat diet (HFD) and Chow groups). We identified differentially expressed genes (DEGs) and module genes using Limma and weighted gene co-expression network analysis (WGCNA), conducted functional enrichment analysis, and employed a machine learning algorithm (least absolute shrinkage and selection operator (LASSO) regression) to pinpoint candidate hub genes for diagnosing obesity-associated risk in offspring of maternal obesity. We constructed a nomogram receiver operating characteristic (ROC) curve to evaluate the diagnostic value. Additionally, we analyzed immune cell infiltration to investigate immune cell dysregulation in maternal obesity. Furthermore, we verified the expression of the candidate hub genes both in vivo and in vitro.
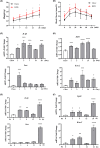
Results: The GSE135830 dataset revealed 2868 DEGs between the mHFD offspring and the mChow group and 2627 WGCNA module genes related to maternal obesity. The overlap of DEGs and module genes in the offspring with maternal obesity in GSE135830 primarily enriched in neurodevelopment and immune regulation. In the GSE127056 dataset, 133 DEGs were identified in the hypothalamus of HFD-induced adult obese individuals. A total of 13 genes intersected between the GSE127056 adult obesity DEGs and the GSE135830 maternal obesity module genes that were primarily enriched in neurodevelopment and the immune response. Following machine learning, two candidate hub genes were chosen for nomogram construction. Diagnostic value evaluation by ROC analysis determined Sytl4 and Kncn2 as hub genes for maternal obesity in the offspring. A gene regulatory network with transcription factor-miRNA interactions was established. Dysregulated immune cells were observed in the hypothalamus of offspring with maternal obesity. Expression of Sytl4 and Kncn2 was validated in a mouse model of hypothalamic inflammation and a palmitic acid-stimulated microglial inflammation model.
Conclusion: Two candidate hub genes (Sytl4 and Kcnc2) were identified and a nomogram was developed to predict obesity risk in offspring with maternal obesity. These findings offer potential diagnostic candidate genes for identifying obesity-associated risks in the offspring of obese mothers.
Keywords: bioinformatics analysis; biomarkers; differentially expressed genes; hypothalamus; obesity.
© 2024 The Authors. CNS Neuroscience & Therapeutics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- BK20211108/Natural Science Foundation of Jiangsu Province
- BK20221274/Natural Science Foundation of Jiangsu Province
- JC2021015/Nantong Science and Technology Project
- M2021106/Scientific Research Project of Health Commission of Jiangsu Province
- JSDW202249/Jiangsu Provincial Medical Key Discipline (Laboratory) Cultivation Unit
- MS2022025/Nantong Municipal Medical Key Laboratory of Molecular Immunology; Nantong Municipal Key Laboratory of Metabolic Immunology and Disease Microenvironment; Scientific Research Project of Health Commission of Nantong
- MSZ2022016/Nantong Municipal Medical Key Laboratory of Molecular Immunology; Nantong Municipal Key Laboratory of Metabolic Immunology and Disease Microenvironment; Scientific Research Project of Health Commission of Nantong
- KD2022KYCXTD005/Scientific Research Innovation Team of Kangda College of Nanjing Medical University
LinkOut - more resources
Full Text Sources

