Enhanced T cell receptor specificity through framework engineering
- PMID: 38545094
- PMCID: PMC10967027
- DOI: 10.3389/fimmu.2024.1345368
Enhanced T cell receptor specificity through framework engineering
Abstract
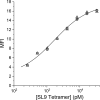
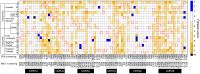
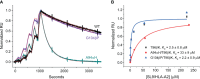
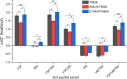
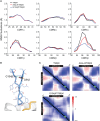
Development of T cell receptors (TCRs) as immunotherapeutics is hindered by inherent TCR cross-reactivity. Engineering more specific TCRs has proven challenging, as unlike antibodies, improving TCR affinity does not usually improve specificity. Although various protein design approaches have been explored to surmount this, mutations in TCR binding interfaces risk broadening specificity or introducing new reactivities. Here we explored if TCR specificity could alternatively be tuned through framework mutations distant from the interface. Studying the 868 TCR specific for the HIV SL9 epitope presented by HLA-A2, we used deep mutational scanning to identify a framework mutation above the mobile CDR3β loop. This glycine to proline mutation had no discernable impact on binding affinity or functional avidity towards the SL9 epitope but weakened recognition of SL9 escape variants and led to fewer responses in a SL9-derived positional scanning library. In contrast, an interfacial mutation near the tip of CDR3α that also did not impact affinity or functional avidity towards SL9 weakened specificity. Simulations indicated that the specificity-enhancing mutation functions by reducing the range of loop motions, limiting the ability of the TCR to adjust to different ligands. Although our results are likely to be TCR dependent, using framework engineering to control TCR loop motions may be a viable strategy for improving the specificity of TCR-based immunotherapies.
Keywords: T cell receptor; framework regions; molecular dynamics; protein engineering; specificity.
Copyright © 2024 Rosenberg, Ayres, Medina-Cucurella, Whitehead and Baker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
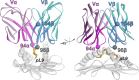

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

