Nanopore-based metagenomics analysis reveals microbial presence in amniotic fluid: A prospective study
- PMID: 38545162
- PMCID: PMC10966708
- DOI: 10.1016/j.heliyon.2024.e28163
Nanopore-based metagenomics analysis reveals microbial presence in amniotic fluid: A prospective study
Abstract
Background: Current research on amniotic fluid (AF) microbiota yields contradictory data, necessitating an accurate, comprehensive, and scientifically rigorous evaluation.
Objective: This study aimed to characterise the microbial features of AF and explore the correlation between microbial information and clinical parameters.
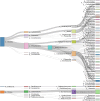
Methods: 76 AF samples were collected in this prospective cohort study. Fourteen samples were utilised to establish the nanopore metagenomic sequencing methodology, whereas the remaining 62 samples underwent a final statistical analysis along with clinical information. Negative controls included the operating room environment (OE), surgical instruments (SI), and laboratory experimental processes (EP) to elucidate the background contamination at each step. Simultaneously, levels of five cytokines (IL-1β, IL-6, IL-8, TNF-α, MMP-8) in AF were assessed.
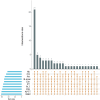
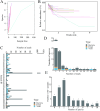
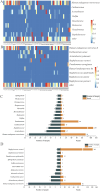
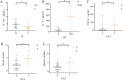
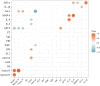
Results: Among the 62 AF samples, microbial analysis identified seven without microbes and 55 with low microbial diversity and abundance. No significant clinical differences were observed between AF samples with and without microbes. The correlation between microbes and clinical parameters in AF with normal chromosomal structure revealed noteworthy findings. In particular, the third trimester exhibited richer microbial diversity. Pseudomonas demonstrated higher detection rates and relative abundance in the second trimester and Preterm Birth (PTB) groups. S. yanoikuyae in the PTB group exhibited elevated detection frequencies and relative abundance. Notably, Pseudomonas negatively correlated with activated partial thromboplastin time (APTT) (r = -0.329, P = 0.016), while Staphylococcus showed positive correlations with APTT (r = 0.395, P = 0.003). Furthermore, Staphylococcus negatively correlated with birth weight (r = -0.297, P = 0.034).
Conclusion: Most AF samples exhibited low microbial diversity and abundance. Certain microbes in AF may correlate with clinical parameters such as gestational age and PTB. However, these associations require further investigation. It is essential to expand the sample size and undertake more comprehensive research to elucidate the clinical implications of microbial presence in AF.
Keywords: Amniotic fluid; Cytokines; Microbes; Nanopore metagenomic sequencing; Pregnancy outcomes.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Lim E.S., Wang D., Holtz L.R. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016;24(10):801–810. - PubMed
LinkOut - more resources
Full Text Sources

