New and Interesting Fungi. 6
- PMID: 38545457
- PMCID: PMC10966675
- DOI: 10.3114/fuse.2023.11.09
New and Interesting Fungi. 6
Abstract
Three new genera, six new species, three combinations, six epitypes, and 25 interesting new host and / or geographical records are introduced in this study. New genera: Neoleptodontidium (based on Neoleptodontidium aquaticum), and Nothoramularia (based on Nothoramularia ragnhildianicola). New species: Acremonium aquaticum (from cooling pad water, USA, Cladophialophora laricicola (on dead wood of Larix sp., Netherlands), Cyphellophora neerlandica (on lichen on brick wall, Netherlands), Geonectria muralis (on moss growing on a wall, Netherlands), Harposporium illinoisense (from rockwool, USA), and Neoleptodontidium aquaticum (from hydroponic water, USA). New combinations: Cyphellophora deltoidea (based on Anthopsis deltoidea), Neoleptodontidium aciculare (based on Leptodontidium aciculare), and Nothoramularia ragnhildianicola (based on Ramularia ragnhildianicola). Epitypes: Cephaliophora tropica (from water, USA), Miricatena prunicola (on leaves of Prunus serotina, Netherlands), Nothoramularia ragnhildianicola (on Ragnhildiana ferruginea, parasitic on Artemisia vulgaris, Germany), Phyllosticta multicorniculata (on needles of Abietis balsamea, Canada), Thyronectria caraganae (on twigs of Caragana arborescens, Ukraine), and Trichosphaeria pilosa (on decayed Salix branch, Netherlands). Furthermore, the higher order phylogeny of three genera regarded as incertae sedis is resolved, namely Cephaliophora (Ascodesmidaceae, Pezizales), Miricatena (Helotiales, Leotiomycetes), and Trichosphaeria (Trichosphaeriaceae, Trichosphaeriales), with Trichosphaeriaceae being an older name for Plectosphaerellaceae. Citation: Crous PW, Akulov A, Balashov S, Boers J, Braun U, Castillo J, Delgado MA, Denman S, Erhard A, Gusella G, Jurjević Ž, Kruse J, Malloch DW, Osieck ER, Polizzi G, Schumacher RK, Slootweg E, Starink-Willemse M, van Iperen AL, Verkley GJM, Groenewald JZ (2023). New and Interesting Fungi. 6. Fungal Systematics and Evolution 11: 109-156. doi: 10.3114/fuse.2023.11.09.
Keywords: ITS barcodes; biodiversity; multi-gene phylogeny; new taxa; systematics; typification.
© 2023 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
Conflict of interest: The authors declare that there is no conflict of interest.
Figures




































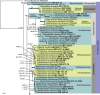


References
-
- Barr ME. (1990). Prodromus to nonlichenized, pyrenomycetous members of class Hymenoascomycetes. Mycotaxon 39: 43–184.
-
- Barr ME, Cannon P. (1994). Discussion 3: Calosphaeriales, Clavicipitales, Coryneliales, Diaporthales, Diatrypales, Halosphaeriales, Hypocreales, Meliolales, Ophiostomatales, Phyllachorales, Sordariales, Trichosphaeriales, and Xylariales. Ascomycete Systematics Problems and Perspectives in the Nineties (Hawksworth DL, ed.) New York & London: Plenum Press: 371–378.
-
- Bell A, Mahoney DP. (1995). Coprophilous fungi from New Zealand. I. Podospora species with swollen agglutinated perithecial hairs. Mycologia 87: 375–396.
LinkOut - more resources
Full Text Sources
