The MUC1-HIF-1α signaling axis regulates pancreatic cancer pathogenesis through polyamine metabolism remodeling
- PMID: 38547055
- PMCID: PMC10998584
- DOI: 10.1073/pnas.2315509121
The MUC1-HIF-1α signaling axis regulates pancreatic cancer pathogenesis through polyamine metabolism remodeling
Abstract
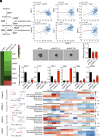
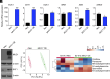
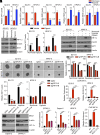
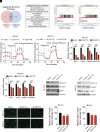
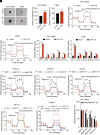
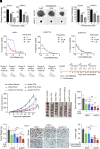
Dysregulation of polyamine metabolism has been implicated in cancer initiation and progression; however, the mechanism of polyamine dysregulation in cancer is not fully understood. In this study, we investigated the role of MUC1, a mucin protein overexpressed in pancreatic cancer, in regulating polyamine metabolism. Utilizing pancreatic cancer patient data, we noted a positive correlation between MUC1 expression and the expression of key polyamine metabolism pathway genes. Functional studies revealed that knockdown of spermidine/spermine N1-acetyltransferase 1 (SAT1), a key enzyme involved in polyamine catabolism, attenuated the oncogenic functions of MUC1, including cell survival and proliferation. We further identified a regulatory axis whereby MUC1 stabilized hypoxia-inducible factor (HIF-1α), leading to increased SAT1 expression, which in turn induced carbon flux into the tricarboxylic acid cycle. MUC1-mediated stabilization of HIF-1α enhanced the promoter occupancy of the latter on SAT1 promoter and corresponding transcriptional activation of SAT1, which could be abrogated by pharmacological inhibition of HIF-1α or CRISPR/Cas9-mediated knockout of HIF1A. MUC1 knockdown caused a significant reduction in the levels of SAT1-generated metabolites, N1-acetylspermidine and N8-acetylspermidine. Given the known role of MUC1 in therapy resistance, we also investigated whether inhibiting SAT1 would enhance the efficacy of FOLFIRINOX chemotherapy. By utilizing organoid and orthotopic pancreatic cancer mouse models, we observed that targeting SAT1 with pentamidine improved the efficacy of FOLFIRINOX, suggesting that the combination may represent a promising therapeutic strategy against pancreatic cancer. This study provides insights into the interplay between MUC1 and polyamine metabolism, offering potential avenues for the development of treatments against pancreatic cancer.
Keywords: MUC1; SAT1; hypoxia-inducible factors; pancreatic cancer; polyamine biosynthesis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Singh P. K., Hollingsworth M. A., Cell surface-associated mucins in signal transduction. Trends Cell Biol. 16, 467–476 (2006). - PubMed
-
- Tsutsumida H., et al. , RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin. Cancer Res. 12, 2976–2987 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

