MIKE: an ultrafast, assembly-, and alignment-free approach for phylogenetic tree construction
- PMID: 38547397
- PMCID: PMC10990684
- DOI: 10.1093/bioinformatics/btae154
MIKE: an ultrafast, assembly-, and alignment-free approach for phylogenetic tree construction
Abstract
Motivation: Constructing a phylogenetic tree requires calculating the evolutionary distance between samples or species via large-scale resequencing data, a process that is both time-consuming and computationally demanding. Striking the right balance between accuracy and efficiency is a significant challenge.
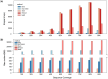
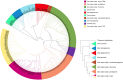
Results: To address this, we introduce a new algorithm, MIKE (MinHash-based k-mer algorithm). This algorithm is designed for the swift calculation of the Jaccard coefficient directly from raw sequencing reads and enables the construction of phylogenetic trees based on the resultant Jaccard coefficient. Simulation results highlight the superior speed of MIKE compared to existing state-of-the-art methods. We used MIKE to reconstruct a phylogenetic tree, incorporating 238 yeast, 303 Zea, 141 Ficus, 67 Oryza, and 43 Saccharum spontaneum samples. MIKE demonstrated accurate performance across varying evolutionary scales, reproductive modes, and ploidy levels, proving itself as a powerful tool for phylogenetic tree construction.
Availability and implementation: MIKE is publicly available on Github at https://github.com/Argonum-Clever2/mike.git.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Batley J, Edwards D.. SNP applications in plants. In Oraguzie NC, Rikkerink EHA, Gardiner SE, De Silva HN (eds) Association Mapping in Plants, pp. 95–102. Berlin: Springer, 2007.
-
- Berlin K, Koren S, Chin C-S. et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 2015;33:623–30. - PubMed
-
- Bos DH, Posada D.. Using models of nucleotide evolution to build phylogenetic trees. Dev Comp Immunol 2005;29:211–27. - PubMed
-
- Buhler J. Efficient large-scale sequence comparison by locality-sensitive hashing. Bioinformatics 2001;17:419–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

