Hypervirulent Klebsiella pneumoniae employs genomic island encoded toxins against bacterial competitors in the gut
- PMID: 38547398
- PMCID: PMC11020217
- DOI: 10.1093/ismejo/wrae054
Hypervirulent Klebsiella pneumoniae employs genomic island encoded toxins against bacterial competitors in the gut
Erratum in
-
Correction to: Hypervirulent Klebsiella pneumoniae employs genomic island encoded toxins against bacterial competitors in the gut.ISME J. 2024 Jan 8;18(1):wrae143. doi: 10.1093/ismejo/wrae143. ISME J. 2024. PMID: 39136613 Free PMC article. No abstract available.
Abstract
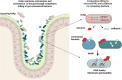
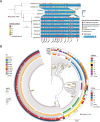
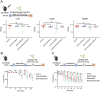
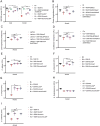
The hypervirulent lineages of Klebsiella pneumoniae (HvKp) cause invasive infections such as Klebsiella-liver abscess. Invasive infection often occurs after initial colonization of the host gastrointestinal tract by HvKp. Over 80% of HvKp isolates belong to the clonal group 23 sublineage I that has acquired genomic islands (GIs) GIE492 and ICEKp10. Our analysis of 12 361 K. pneumoniae genomes revealed that GIs GIE492 and ICEKp10 are co-associated with the CG23-I and CG10118 HvKp lineages. GIE492 and ICEKp10 enable HvKp to make a functional bacteriocin microcin E492 (mccE492) and the genotoxin colibactin, respectively. We discovered that GIE492 and ICEKp10 play cooperative roles and enhance gastrointestinal colonization by HvKp. Colibactin is the primary driver of this effect, modifying gut microbiome diversity. Our in vitro assays demonstrate that colibactin and mccE492 kill or inhibit a range of Gram-negative Klebsiella species and Escherichia coli strains, including Gram-positive bacteria, sometimes cooperatively. Moreover, mccE492 and colibactin kill human anaerobic gut commensals that are similar to the taxa found altered by colibactin in the mouse intestines. Our findings suggest that GIs GIE492 and ICEKp10 enable HvKp to kill several commensal bacterial taxa during interspecies interactions in the gut. Thus, acquisition of GIE492 and ICEKp10 could enable better carriage in host populations and explain the dominance of the CG23-I HvKp lineage.
Keywords: Klebsiella pneumoniae; colibactin; colonization; commensal; genomic island; gut; hypervirulent; microcin.
© The Author(s) [2024]. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
We declare that the authors do not possess any competing financial interests in relation to the work published.
Figures





References
-
- Belk WP. Pulmonary infections by Friedländer’s bacillus. J Infect Dis 1926;38:115–26. 10.1093/infdis/38.2.115 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

