A personalized medicine approach identifies enasidenib as an efficient treatment for IDH2 mutant chondrosarcoma
- PMID: 38547578
- PMCID: PMC10990714
- DOI: 10.1016/j.ebiom.2024.105090
A personalized medicine approach identifies enasidenib as an efficient treatment for IDH2 mutant chondrosarcoma
Abstract
Background: Sarcomas represent an extensive group of malignant diseases affecting mesodermal tissues. Among sarcomas, the clinical management of chondrosarcomas remains a complex challenge, as high-grade tumours do not respond to current therapies. Mutations in the isocitrate dehydrogenase (IDH) 1 and 2 genes are among the most common mutations detected in chondrosarcomas and may represent a therapeutic opportunity. The presence of mutated IDH (mIDH) enzymes results in the accumulation of the oncometabolite 2-HG leading to molecular alterations that contribute to drive tumour growth.
Methods: We developed a personalized medicine strategy based on the targeted NGS/Sanger sequencing of sarcoma samples (n = 6) and the use of matched patient-derived cell lines as a drug-testing platform. The anti-tumour potential of IDH mutations found in two chondrosarcoma cases was analysed in vitro, in vivo and molecularly (transcriptomic and DNA methylation analyses).
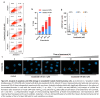
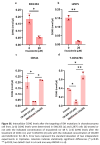
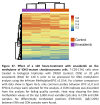
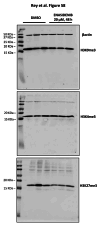
Findings: We treated several chondrosarcoma models with specific mIDH1/2 inhibitors. Among these treatments, only the mIDH2 inhibitor enasidenib was able to decrease 2-HG levels and efficiently reduce the viability of mIDH2 chondrosarcoma cells. Importantly, oral administration of enasidenib in xenografted mice resulted in a complete abrogation of tumour growth. Enasidenib induced a profound remodelling of the transcriptomic landscape not associated to changes in the 5 mC methylation levels and its anti-tumour effects were associated with the repression of proliferative pathways such as those controlled by E2F factors.
Interpretation: Overall, this work provides preclinical evidence for the use of enasidenib to treat mIDH2 chondrosarcomas.
Funding: Supported by the Spanish Research Agency/FEDER (grants PID2022-142020OB-I00; PID2019-106666RB-I00), the ISC III/FEDER (PI20CIII/00020; DTS18CIII/00005; CB16/12/00390; CB06/07/1009; CB19/07/00057); the GEIS group (GEIS-62); and the PCTI (Asturias)/FEDER (IDI/2021/000027).
Keywords: Chondrosarcoma; Enasidenib; IDH2; Patient-derived models; Personalized medicine.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no conflict of interest.
Figures









References
-
- Sayles L.C., Breese M.R., Koehne A.L., et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63. doi: 10.1158/2159-8290.CD-17-1152. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

