Automatic categorization of self-acknowledged limitations in randomized controlled trial publications
- PMID: 38548008
- PMCID: PMC11807350
- DOI: 10.1016/j.jbi.2024.104628
Automatic categorization of self-acknowledged limitations in randomized controlled trial publications
Abstract
Objective: Acknowledging study limitations in a scientific publication is a crucial element in scientific transparency and progress. However, limitation reporting is often inadequate. Natural language processing (NLP) methods could support automated reporting checks, improving research transparency. In this study, our objective was to develop a dataset and NLP methods to detect and categorize self-acknowledged limitations (e.g., sample size, blinding) reported in randomized controlled trial (RCT) publications.
Methods: We created a data model of limitation types in RCT studies and annotated a corpus of 200 full-text RCT publications using this data model. We fine-tuned BERT-based sentence classification models to recognize the limitation sentences and their types. To address the small size of the annotated corpus, we experimented with data augmentation approaches, including Easy Data Augmentation (EDA) and Prompt-Based Data Augmentation (PromDA). We applied the best-performing model to a set of about 12K RCT publications to characterize self-acknowledged limitations at larger scale.
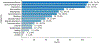
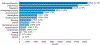
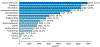
Results: Our data model consists of 15 categories and 24 sub-categories (e.g., Population and its sub-category DiagnosticCriteria). We annotated 1090 instances of limitation types in 952 sentences (4.8 limitation sentences and 5.5 limitation types per article). A fine-tuned PubMedBERT model for limitation sentence classification improved upon our earlier model by about 1.5 absolute percentage points in F1 score (0.821 vs. 0.8) with statistical significance (p<.001). Our best-performing limitation type classification model, PubMedBERT fine-tuning with PromDA (Output View), achieved an F1 score of 0.7, improving upon the vanilla PubMedBERT model by 2.7 percentage points, with statistical significance (p<.001).
Conclusion: The model could support automated screening tools which can be used by journals to draw the authors' attention to reporting issues. Automatic extraction of limitations from RCT publications could benefit peer review and evidence synthesis, and support advanced methods to search and aggregate the evidence from the clinical trial literature.
Keywords: Large language models; Natural language processing; Randomized controlled trials; Reporting quality; Self-acknowledged limitations; Text classification.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Halil Kilicoglu reports financial support was provided by National Library of Medicine.
Figures



Similar articles
-
SPIRIT-CONSORT-TM: a corpus for assessing transparency of clinical trial protocol and results publications.Sci Data. 2025 Feb 28;12(1):355. doi: 10.1038/s41597-025-04629-1. Sci Data. 2025. PMID: 40021657 Free PMC article.
-
Text classification models for assessing the completeness of randomized controlled trial publications based on CONSORT reporting guidelines.Sci Rep. 2024 Sep 17;14(1):21721. doi: 10.1038/s41598-024-72130-7. Sci Rep. 2024. PMID: 39289403 Free PMC article.
-
CONSORT-TM: Text classification models for assessing the completeness of randomized controlled trial publications.medRxiv [Preprint]. 2024 Apr 1:2024.03.31.24305138. doi: 10.1101/2024.03.31.24305138. medRxiv. 2024. Update in: Sci Rep. 2024 Sep 17;14(1):21721. doi: 10.1038/s41598-024-72130-7. PMID: 38633775 Free PMC article. Updated. Preprint.
-
Sample size, study length, and inadequate controls were the most common self-acknowledged limitations in manual therapy trials: A methodological review.J Clin Epidemiol. 2021 Feb;130:96-106. doi: 10.1016/j.jclinepi.2020.10.018. Epub 2020 Nov 1. J Clin Epidemiol. 2021. PMID: 33144246 Review.
-
Natural Language Processing Applications in the Clinical Neurosciences: A Machine Learning Augmented Systematic Review.Acta Neurochir Suppl. 2022;134:277-289. doi: 10.1007/978-3-030-85292-4_32. Acta Neurochir Suppl. 2022. PMID: 34862552
Cited by
-
SPIRIT-CONSORT-TM: a corpus for assessing transparency of clinical trial protocol and results publications.Sci Data. 2025 Feb 28;12(1):355. doi: 10.1038/s41597-025-04629-1. Sci Data. 2025. PMID: 40021657 Free PMC article.
-
SPIRIT-CONSORT-TM: a corpus for assessing transparency of clinical trial protocol and results publications.medRxiv [Preprint]. 2025 Jan 15:2025.01.14.25320543. doi: 10.1101/2025.01.14.25320543. medRxiv. 2025. Update in: Sci Data. 2025 Feb 28;12(1):355. doi: 10.1038/s41597-025-04629-1. PMID: 39867389 Free PMC article. Updated. Preprint.
-
The emergence of large language models as tools in literature reviews: a large language model-assisted systematic review.J Am Med Inform Assoc. 2025 Jun 1;32(6):1071-1086. doi: 10.1093/jamia/ocaf063. J Am Med Inform Assoc. 2025. PMID: 40332983 Free PMC article.
References
-
- Else H, How a torrent of COVID science changed research publishing-in seven charts, Nature (2020) 553. - PubMed
-
- Watson C, Rise of the preprint: how rapid data sharing during COVID-19 has changed science forever, Nat. Med 28 (1) (2022) 2–5. - PubMed
-
- Bramstedt KA, The carnage of substandard research during the COVID-19 pandemic: a call for quality, J. Med. Ethics 46 (12) (2020) 803–807. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

