Structure and function of Semaphorin-5A glycosaminoglycan interactions
- PMID: 38548715
- PMCID: PMC10978931
- DOI: 10.1038/s41467-024-46725-7
Structure and function of Semaphorin-5A glycosaminoglycan interactions
Abstract
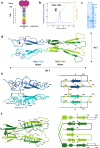
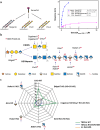
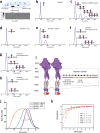
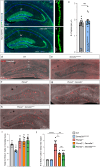
Integration of extracellular signals by neurons is pivotal for brain development, plasticity, and repair. Axon guidance relies on receptor-ligand interactions crosstalking with extracellular matrix components. Semaphorin-5A (Sema5A) is a bifunctional guidance cue exerting attractive and inhibitory effects on neuronal growth through the interaction with heparan sulfate (HS) and chondroitin sulfate (CS) glycosaminoglycans (GAGs), respectively. Sema5A harbors seven thrombospondin type-1 repeats (TSR1-7) important for GAG binding, however the underlying molecular basis and functions in vivo remain enigmatic. Here we dissect the structural basis for Sema5A:GAG specificity and demonstrate the functional significance of this interaction in vivo. Using x-ray crystallography, we reveal a dimeric fold variation for TSR4 that accommodates GAG interactions. TSR4 co-crystal structures identify binding residues validated by site-directed mutagenesis. In vitro and cell-based assays uncover specific GAG epitopes necessary for TSR association. We demonstrate that HS-GAG binding is preferred over CS-GAG and mediates Sema5A oligomerization. In vivo, Sema5A:GAG interactions are necessary for Sema5A function and regulate Plexin-A2 dependent dentate progenitor cell migration. Our study rationalizes Sema5A associated developmental and neurological disorders and provides mechanistic insights into how multifaceted guidance functions of a single transmembrane cue are regulated by proteoglycans.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

