Autophagy-enhancing ATG16L1 polymorphism is associated with improved clinical outcome and T-cell immunity in chronic HIV-1 infection
- PMID: 38548722
- PMCID: PMC10979031
- DOI: 10.1038/s41467-024-46606-z
Autophagy-enhancing ATG16L1 polymorphism is associated with improved clinical outcome and T-cell immunity in chronic HIV-1 infection
Abstract
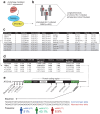
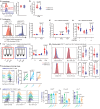
Chronic HIV-1 infection is characterized by T-cell dysregulation that is partly restored by antiretroviral therapy. Autophagy is a critical regulator of T-cell function. Here, we demonstrate a protective role for autophagy in HIV-1 disease pathogenesis. Targeted analysis of genetic variation in core autophagy gene ATG16L1 reveals the previously unidentified rs6861 polymorphism, which correlates functionally with enhanced autophagy and clinically with improved survival of untreated HIV-1-infected individuals. T-cells carrying ATG16L1 rs6861(TT) genotype display improved antiviral immunity, evidenced by increased proliferation, revamped immune responsiveness, and suppressed exhaustion/immunosenescence features. In-depth flow-cytometric and transcriptional profiling reveal T-helper-cell-signatures unique to rs6861(TT) individuals with enriched regulation of pro-inflammatory networks and skewing towards immunoregulatory phenotype. Therapeutic enhancement of autophagy recapitulates the rs6861(TT)-associated T-cell traits in non-carriers. These data underscore the in vivo relevance of autophagy for longer-lasting T-cell-mediated HIV-1 control, with implications towards development of host-directed antivirals targeting autophagy to restore immune function in chronic HIV-1 infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

