A midbrain GABAergic circuit constrains wakefulness in a mouse model of stress
- PMID: 38548744
- PMCID: PMC10978901
- DOI: 10.1038/s41467-024-46707-9
A midbrain GABAergic circuit constrains wakefulness in a mouse model of stress
Abstract
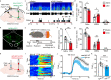
Enhancement of wakefulness is a prerequisite for adaptive behaviors to cope with acute stress, but hyperarousal is associated with impaired behavioral performance. Although the neural circuitries promoting wakefulness in acute stress conditions have been extensively identified, less is known about the circuit mechanisms constraining wakefulness to prevent hyperarousal. Here, we found that chemogenetic or optogenetic activation of GAD2-positive GABAergic neurons in the midbrain dorsal raphe nucleus (DRNGAD2) decreased wakefulness, while inhibition or ablation of these neurons produced an increase in wakefulness along with hyperactivity. Surprisingly, DRNGAD2 neurons were paradoxically wakefulness-active and were further activated by acute stress. Bidirectional manipulations revealed that DRNGAD2 neurons constrained the increase of wakefulness and arousal level in a mouse model of stress. Circuit-specific investigations demonstrated that DRNGAD2 neurons constrained wakefulness via inhibition of the wakefulness-promoting paraventricular thalamus. Therefore, the present study identified a wakefulness-constraining role DRNGAD2 neurons in acute stress conditions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

