Macrophage migration inhibitory factor blockade reprograms macrophages and disrupts prosurvival signaling in acute myeloid leukemia
- PMID: 38548753
- PMCID: PMC10978870
- DOI: 10.1038/s41420-024-01924-5
Macrophage migration inhibitory factor blockade reprograms macrophages and disrupts prosurvival signaling in acute myeloid leukemia
Abstract
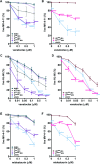
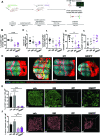
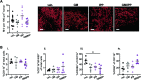
The malignant microenvironment plays a major role in the development of resistance to therapies and the occurrence of relapses in acute myeloid leukemia (AML). We previously showed that interactions of AML blasts with bone marrow macrophages (MΦ) shift their polarization towards a protumoral (M2-like) phenotype, promoting drug resistance; we demonstrated that inhibiting the colony-stimulating factor-1 receptor (CSF1R) repolarizes MΦ towards an antitumoral (M1-like) phenotype and that other factors may be involved. We investigated here macrophage migration inhibitory factor (MIF) as a target in AML blast survival and protumoral interactions with MΦ. We show that pharmacologically inhibiting MIF secreted by AML blasts results in their apoptosis. However, this effect is abrogated when blasts are co-cultured in close contact with M2-like MΦ. We next demonstrate that pharmacological inhibition of MIF secreted by MΦ, in the presence of granulocyte macrophage-colony stimulating factor (GM-CSF), efficiently reprograms MΦ to an M1-like phenotype that triggers apoptosis of interacting blasts. Furthermore, contact with reprogrammed MΦ relieves blast resistance to venetoclax and midostaurin acquired in contact with CD163+ protumoral MΦ. Using intravital imaging in mice, we also show that treatment with MIF inhibitor 4-IPP and GM-CSF profoundly affects the tumor microenvironment in vivo: it strikingly inhibits tumor vasculature, reduces protumoral MΦ, and slows down leukemia progression. Thus, our data demonstrate that MIF plays a crucial role in AML MΦ M2-like protumoral phenotype that can be reversed by inhibiting its activity and suggest the therapeutic targeting of MIF as an avenue towards improved AML treatment outcomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

