A multiparameter liquid biopsy approach allows to track melanoma dynamics and identify early treatment resistance
- PMID: 38548846
- PMCID: PMC10978909
- DOI: 10.1038/s41698-024-00567-0
A multiparameter liquid biopsy approach allows to track melanoma dynamics and identify early treatment resistance
Abstract
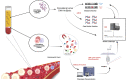
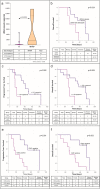
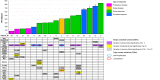
Melanoma heterogeneity is a hurdle in metastatic disease management. Although the advent of targeted therapy has significantly improved patient outcomes, the occurrence of resistance makes monitoring of the tumor genetic landscape mandatory. Liquid biopsy could represent an important biomarker for the real-time tracing of disease evolution. Thus, we aimed to correlate liquid biopsy dynamics with treatment response and progression by devising a multiplatform approach applied to longitudinal melanoma patient monitoring. We conceived an approach that exploits Next Generation Sequencing (NGS) and droplet digital PCR, as well as the FDA-cleared platform CellSearch, to analyze circulating tumor DNA (ctDNA) trend and circulating melanoma cell (CMC) count, together with their customized genetic and copy number variation analysis. The approach was applied to 17 stage IV melanoma patients treated with BRAF/MEK inhibitors, followed for up to 28 months. BRAF mutations were detected in the plasma of 82% of patients. Single nucleotide variants known or suspected to confer resistance were identified in 70% of patients. Moreover, the amount of ctDNA, both at baseline and during response, correlated with the type and duration of the response itself, and the CMC count was confirmed to be a prognostic biomarker. This work provides proof of principle of the power of this approach and paves the way for a validation study aimed at evaluating early ctDNA-guided treatment decisions in stage IV melanoma. The NGS-based molecular profile complemented the analysis of ctDNA trend and, together with CMC analysis, revealed to be useful in capturing tumor evolution.
© 2024. The Author(s).
Conflict of interest statement
MCS received a travel grant from Agilent Technologies, all the other authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

