High prevalence of carbapenem resistance and clonal expansion of blaNDM gene in Klebsiella pneumoniae isolates in an Iranian referral pediatric hospital
- PMID: 38549114
- PMCID: PMC10976756
- DOI: 10.1186/s13099-024-00611-1
High prevalence of carbapenem resistance and clonal expansion of blaNDM gene in Klebsiella pneumoniae isolates in an Iranian referral pediatric hospital
Abstract
Background: The increasing global concern regarding antibiotic resistance necessitates in-depth studies to comprehend the phenotypic and genotypic characteristics of resistant bacterial strains. This study aimed to investigate the prevalence, antibiotic resistance profiles, and molecular characteristics of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates in an Iranian referral pediatrics hospital.
Methods: In this study, we examined CRKP isolates collected from hospitalized pediatric patients across various wards. The isolates underwent antimicrobial susceptibility testing, the polymerase chain reaction (PCR) analysis for carbapenemase genes (blaNDM, blaVIM and blaIMP), and genetic relatedness assessment using pulsed-field gel electrophoresis (PFGE).
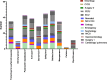
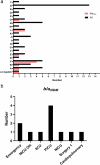
Results: Among 166 K. pneumoniae isolates, 54 (32.5%) exhibited resistance to carbapenems. Notably, all these resistant isolates were resistant to imipenem, with 35 (65%) displaying resistance to both imipenem and meropenem. Of the 54 CRKP isolates, 24 (44%) were metallo-β-lactamases (MBL)-producing. The prevalence of the blaNDM gene among CKCP and MBL-producing isolates was 20% (n = 11) and 44% (n = 24), respectively. The blaVIM and blaIMP genes were not detected in any of the isolates. Twenty-six CRKP isolates (48%) were recovered from ICUs. PFGE analysis of CRKP isolates revealed 20 clusters, with cluster S being the most prevalent, comprising 24% of the total (n = 13).
Conclusion: Our study reveals a concerning prevalence of carbapenem resistance in K. pneumoniae isolates. Specifically, the detection of the blaNDM gene in 20% of CRKP isolates, with a significant proportion (82%) observed in isolated CRKP from the ICUs and emergency departments, underscores the potential clonal expansion of these resistant strains within these critical hospital wards.
Keywords: Klebsiella pneumoniae; bla NDM; Carbapenem resistance; Genotyping.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Resistance Phenotype and Molecular Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae Isolates in Shanghai.Microb Drug Resist. 2021 Oct;27(10):1312-1318. doi: 10.1089/mdr.2020.0390. Epub 2021 Jul 23. Microb Drug Resist. 2021. PMID: 34297609
-
Isolation of Hv-CRKP with co-production of three carbapenemases (blaKPC, blaOXA-181 or OXA-232, and blaNDM-1) and a virulence plasmid: a study from a Chinese tertiary hospital.Front Microbiol. 2023 May 24;14:1182870. doi: 10.3389/fmicb.2023.1182870. eCollection 2023. Front Microbiol. 2023. PMID: 37293218 Free PMC article.
-
[Comparison of phenotypic methods and polymerase chain reaction for the detection of carbapenemase production in clinical Klebsiella pneumoniae isolates].Mikrobiyol Bul. 2017 Jul;51(3):269-276. doi: 10.5578/mb.57333. Mikrobiyol Bul. 2017. PMID: 28929963 Turkish.
-
Analysis of carbapenemases genes of carbapenem-resistant Klebsiella pneumoniae isolated from Tehran heart center.Iran J Microbiol. 2022 Feb;14(1):38-46. doi: 10.18502/ijm.v14i1.8799. Iran J Microbiol. 2022. PMID: 35664723 Free PMC article.
-
Carbapenem-resistant hypermucoviscous Klebsiella pneumoniae clinical isolates from a tertiary hospital in China: Antimicrobial susceptibility, resistance phenotype, epidemiological characteristics, microbial virulence, and risk factors.Front Cell Infect Microbiol. 2022 Dec 21;12:1083009. doi: 10.3389/fcimb.2022.1083009. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36619764 Free PMC article.
Cited by
-
Identification of beta-lactamase genes and molecular genotyping of multidrug-resistant clinical isolates of Klebsiella pneumoniae.BMC Microbiol. 2024 Dec 28;24(1):549. doi: 10.1186/s12866-024-03679-6. BMC Microbiol. 2024. PMID: 39732628 Free PMC article.
-
Molecular detection of OXA-48 and NDM-1 carbapenemase genes among clinical isolates of Klebsiella pneumoniae recovered from patients attending a private tertiary hospital in Southwestern Nigeria.BMC Infect Dis. 2024 Sep 13;24(1):970. doi: 10.1186/s12879-024-09869-x. BMC Infect Dis. 2024. PMID: 39271986 Free PMC article.
-
New insights and perspectives on the virulence of hypervirulent Klebsiella pneumoniae.Folia Microbiol (Praha). 2025 Jun;70(3):517-533. doi: 10.1007/s12223-025-01261-9. Epub 2025 Apr 8. Folia Microbiol (Praha). 2025. PMID: 40198504 Review.
References
-
- Mamishi S, Mahmoudi S, Naserzadeh N, Hosseinpour Sadeghi R, Haghi Ashtiani MT, Bahador A, et al. Antibiotic resistance and genotyping of gram-negative bacteria causing hospital-acquired infection in patients referred to children’s Medical Center. Infect Drug Resist. 2019;12:3377–84. doi: 10.2147/IDR.S195126. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

