A divergent protein kinase A regulatory subunit essential for morphogenesis of the human pathogen Leishmania
- PMID: 38551993
- PMCID: PMC11006142
- DOI: 10.1371/journal.ppat.1012073
A divergent protein kinase A regulatory subunit essential for morphogenesis of the human pathogen Leishmania
Abstract
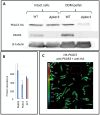
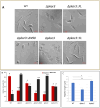
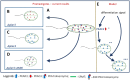
Parasitic protozoa of the genus Leishmania cycle between the phagolysosome of mammalian macrophages, where they reside as rounded intracellular amastigotes, and the midgut of female sand flies, which they colonize as elongated extracellular promastigotes. Previous studies indicated that protein kinase A (PKA) plays an important role in the initial steps of promastigote differentiation into amastigotes. Here, we describe a novel regulatory subunit of PKA (which we have named PKAR3) that is unique to Leishmania and most (but not all) other Kinetoplastidae. PKAR3 is localized to subpellicular microtubules (SPMT) in the cell cortex, where it recruits a specific catalytic subunit (PKAC3). Promastigotes of pkar3 or pkac3 null mutants lose their elongated shape and become rounded but remain flagellated. Truncation of an N-terminal formin homology (FH)-like domain of PKAR3 results in its detachment from the SPMT, also leading to rounded promastigotes. Thus, the tethering of PKAC3 via PKAR3 at the cell cortex is essential for maintenance of the elongated shape of promastigotes. This role of PKAR3 is reminiscent of PKARIβ and PKARIIβ binding to microtubules of mammalian neurons, which is essential for the elongation of dendrites and axons, respectively. Interestingly, PKAR3 binds nucleoside analogs, but not cAMP, with a high affinity similar to the PKAR1 isoform of Trypanosoma. We propose that these early-diverged protists have re-purposed PKA for a novel signaling pathway that spatiotemporally controls microtubule remodeling and cell shape.
Copyright: © 2024 Fischer Weinberger et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Maxfield L, Crane JS, Leishmaniasis (StatPearls Publishing, 2020). - PubMed
-
- Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol Biochem Parasitol. 2005;141: 99–108. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

