A bioinformatics approach to elucidate conserved genes and pathways in C. elegans as an animal model for cardiovascular research
- PMID: 38553458
- PMCID: PMC10980734
- DOI: 10.1038/s41598-024-56562-9
A bioinformatics approach to elucidate conserved genes and pathways in C. elegans as an animal model for cardiovascular research
Abstract
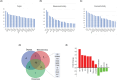
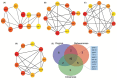
Cardiovascular disease (CVD) is a collective term for disorders of the heart and blood vessels. The molecular events and biochemical pathways associated with CVD are difficult to study in clinical settings on patients and in vitro conditions. Animal models play a pivotal and indispensable role in CVD research. Caenorhabditis elegans, a nematode species, has emerged as a prominent experimental organism widely utilized in various biomedical research fields. However, the specific number of CVD-related genes and pathways within the C. elegans genome remains undisclosed to date, limiting its in-depth utilization for investigations. In the present study, we conducted a comprehensive analysis of genes and pathways related to CVD within the genomes of humans and C. elegans through a systematic bioinformatic approach. A total of 1113 genes in C. elegans orthologous to the most significant CVD-related genes in humans were identified, and the GO terms and pathways were compared to study the pathways that are conserved between the two species. In order to infer the functions of CVD-related orthologous genes in C. elegans, a PPI network was constructed. Orthologous gene PPI network analysis results reveal the hubs and important KRs: pmk-1, daf-21, gpb-1, crh-1, enpl-1, eef-1G, acdh-8, hif-1, pmk-2, and aha-1 in C. elegans. Modules were identified for determining the role of the orthologous genes at various levels in the created network. We also identified 9 commonly enriched pathways between humans and C. elegans linked with CVDs that include autophagy (animal), the ErbB signaling pathway, the FoxO signaling pathway, the MAPK signaling pathway, ABC transporters, the biosynthesis of unsaturated fatty acids, fatty acid metabolism, glutathione metabolism, and metabolic pathways. This study provides the first systematic genomic approach to explore the CVD-associated genes and pathways that are present in C. elegans, supporting the use of C. elegans as a prominent animal model organism for cardiovascular diseases.
Keywords: C. elegans; Animal models; Cardiovascular disease; Orthologous genes; PPI network.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Conserved Cardiovascular Network: Bioinformatics Insights into Genes and Pathways for Establishing Caenorhabditis elegans as an Animal Model for Cardiovascular Diseases.bioRxiv [Preprint]. 2024 Jan 7:2023.12.24.573256. doi: 10.1101/2023.12.24.573256. bioRxiv. 2024. PMID: 38234826 Free PMC article. Preprint.
-
PMK-1 p38 MAPK promotes cadmium stress resistance, the expression of SKN-1/Nrf and DAF-16 target genes, and protein biosynthesis in Caenorhabditis elegans.Mol Genet Genomics. 2017 Dec;292(6):1341-1361. doi: 10.1007/s00438-017-1351-z. Epub 2017 Aug 1. Mol Genet Genomics. 2017. PMID: 28766017 Free PMC article.
-
Wide diversity in structure and expression profiles among members of the Caenorhabditis elegans globin protein family.BMC Genomics. 2007 Oct 4;8:356. doi: 10.1186/1471-2164-8-356. BMC Genomics. 2007. PMID: 17916248 Free PMC article.
-
DAF-16: FOXO in the Context of C. elegans.Curr Top Dev Biol. 2018;127:1-21. doi: 10.1016/bs.ctdb.2017.11.007. Epub 2018 Feb 2. Curr Top Dev Biol. 2018. PMID: 29433733 Review.
-
TGF-beta signaling.WormBook. 2005 Sep 9:1-12. doi: 10.1895/wormbook.1.22.1. WormBook. 2005. PMID: 18050404 Free PMC article. Review.
Cited by
-
Unveiling the Mechanisms Underlying the Immunotherapeutic Potential of Gene-miRNA and Drugs in Head and Neck Cancer.Pharmaceuticals (Basel). 2024 Jul 10;17(7):921. doi: 10.3390/ph17070921. Pharmaceuticals (Basel). 2024. PMID: 39065771 Free PMC article.
-
Label-Free Proteomic Profiling of the dvls2 (CL2006) Caenorhabditis elegans Alzheimer's Disease (AD) Model Reveals Conserved Molecular Signatures Shared With the Human AD Brain.J Neurochem. 2025 Jul;169(7):e70152. doi: 10.1111/jnc.70152. J Neurochem. 2025. PMID: 40653809 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

