Identification of spatially-resolved markers of malignant transformation in Intraductal Papillary Mucinous Neoplasms
- PMID: 38553466
- PMCID: PMC10980816
- DOI: 10.1038/s41467-024-46994-2
Identification of spatially-resolved markers of malignant transformation in Intraductal Papillary Mucinous Neoplasms
Abstract
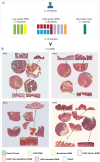
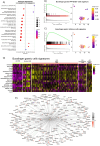
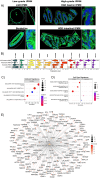
The existing Intraductal Papillary Mucinous Neoplasm (IPMN) risk stratification relies on clinical and histological factors, resulting in inaccuracies and leading to suboptimal treatment. This is due to the lack of appropriate molecular markers that can guide patients toward the best therapeutic options. Here, we assess and confirm subtype-specific markers for IPMN across two independent cohorts of patients using two Spatial Transcriptomics (ST) technologies. Specifically, we identify HOXB3 and ZNF117 as markers for Low-Grade Dysplasia, SPDEF and gastric neck cell markers in borderline cases, and NKX6-2 and gastric isthmus cell markers in High-Grade-Dysplasia Gastric IPMN, highlighting the role of TNFα and MYC activation in IPMN progression and the role of NKX6-2 in the specific Gastric IPMN progression. In conclusion, our work provides a step forward in understanding the gene expression landscapes of IPMN and the critical transcriptional networks related to PDAC progression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

