Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont
- PMID: 38553475
- PMCID: PMC10980776
- DOI: 10.1038/s41522-024-00503-x
Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont
Erratum in
-
Publisher Correction: Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont.NPJ Biofilms Microbiomes. 2024 May 3;10(1):43. doi: 10.1038/s41522-024-00515-7. NPJ Biofilms Microbiomes. 2024. PMID: 38702335 Free PMC article. No abstract available.
Abstract
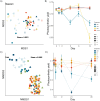
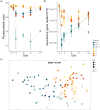
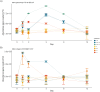
Host-associated microbiota are critical for eukaryotic host functioning, to the extent that hosts and their associated microbial communities are often considered "holobionts". Most studies of holobionts have focused on descriptive approaches or have used model systems, usually in the laboratory, to understand host-microbiome interactions. To advance our understanding of host-microbiota interactions and their wider ecological impacts, we need experimental frameworks that can explore causation in non-model hosts, which often have highly diverse microbiota, and in their natural ecological setting (i.e. in the field). We used a dominant habitat-forming seaweed, Hormosira banksii, to explore these issues and to experimentally test host-microbiota interactions in a non-model holobiont. The experimental protocols were aimed at trying to disentangle microbially mediated effects on hosts from direct effects on hosts associated with the methods employed to manipulate host-microbiota. This was done by disrupting the microbiome, either through removal/disruption using a combination of antimicrobial treatments, or additions of specific taxa via inoculations, or a combination of thew two. The experiments were done in mesocosms and in the field. Three different antibiotic treatments were used to disrupt seaweed-associated microbiota to test whether disturbances of microbiota, particularly bacteria, would negatively affect host performance. Responses of bacteria to these disturbances were complex and differed substantially among treatments, with some antibacterial treatments having little discernible effect. However, the temporal sequence of responses antibiotic treatments, changes in bacterial diversity and subsequent decreases in host performance, strongly suggested an effect of the microbiota on host performance in some treatments, as opposed to direct effects of the antibiotics. To further test these effects, we used 16S-rRNA-gene sequencing to identify bacterial taxa that were either correlated, or uncorrelated, with poor host performance following antibiotic treatment. These were then isolated and used in inoculation experiments, independently or in combination with the previously used antibiotic treatments. Negative effects on host performance were strongest where specific microbial antimicrobials treatments were combined with inoculations of strains that were correlated with poor host performance. For these treatments, negative host effects persisted the entire experimental period (12 days), even though treatments were only applied at the beginning of the experiment. Host performance recovered in all other treatments. These experiments provide a framework for exploring causation and disentangling microbially mediated vs. direct effects on hosts for ecologically important, non-model holobionts in the field. This should allow for better predictions of how these systems will respond to, and potentially mitigate, environmental disturbances in their natural context.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
- Holsworth Wildlife Endowment/Ecological Society of Australia (ESA)
- DP180104041/Department of Education and Training | Australian Research Council (ARC)
- DP180104041/Department of Education and Training | Australian Research Council (ARC)
- DP180104041/Department of Education and Training | Australian Research Council (ARC)
LinkOut - more resources
Full Text Sources

