Insights on E1-like enzyme ATG7: functional regulation and relationships with aging-related diseases
- PMID: 38553562
- PMCID: PMC10980737
- DOI: 10.1038/s42003-024-06080-1
Insights on E1-like enzyme ATG7: functional regulation and relationships with aging-related diseases
Abstract
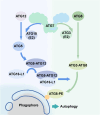
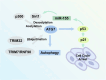
Autophagy is a dynamic self-renovation biological process that maintains cell homeostasis and is responsible for the quality control of proteins, organelles, and energy metabolism. The E1-like ubiquitin-activating enzyme autophagy-related gene 7 (ATG7) is a critical factor that initiates classic autophagy reactions by promoting the formation and extension of autophagosome membranes. Recent studies have identified the key functions of ATG7 in regulating the cell cycle, apoptosis, and metabolism associated with the occurrence and development of multiple diseases. This review summarizes how ATG7 is precisely programmed by genetic, transcriptional, and epigenetic modifications in cells and the relationship between ATG7 and aging-related diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The FAP motif within human ATG7, an autophagy-related E1-like enzyme, is essential for the E2-substrate reaction of LC3 lipidation.Autophagy. 2012 Jan;8(1):88-97. doi: 10.4161/auto.8.1.18339. Epub 2012 Jan 1. Autophagy. 2012. PMID: 22170151
-
Structures of Atg7-Atg3 and Atg7-Atg10 reveal noncanonical mechanisms of E2 recruitment by the autophagy E1.Autophagy. 2013 May;9(5):778-80. doi: 10.4161/auto.23644. Epub 2013 Feb 6. Autophagy. 2013. PMID: 23388412 Free PMC article.
-
Purification and characterization of a ubiquitin-like system for autophagosome formation.J Microbiol Biotechnol. 2010 Dec;20(12):1647-52. J Microbiol Biotechnol. 2010. PMID: 21193819
-
Allosteric regulation through a switch element in the autophagy E2, Atg3.Autophagy. 2020 Jan;16(1):183-184. doi: 10.1080/15548627.2019.1688550. Epub 2019 Nov 9. Autophagy. 2020. PMID: 31690182 Free PMC article. Review.
-
In vitro assays of lipidation of Mammalian Atg8 homologs.Curr Protoc Cell Biol. 2014 Sep 2;64:11.20.1-13. doi: 10.1002/0471143030.cb1120s64. Curr Protoc Cell Biol. 2014. PMID: 25181299 Review.
Cited by
-
Canonical and noncanonical autophagy: involvement in Parkinson's disease.Front Cell Dev Biol. 2025 Jan 30;13:1518991. doi: 10.3389/fcell.2025.1518991. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39949604 Free PMC article. Review.
-
Enhanced secretion of the amyotrophic lateral sclerosis ALS-associated misfolded TDP-43 mediated by the ER-ubiquitin specific peptidase USP19.Cell Mol Life Sci. 2025 Feb 13;82(1):76. doi: 10.1007/s00018-025-05589-w. Cell Mol Life Sci. 2025. PMID: 39948244 Free PMC article.
-
Advancing muscle aging and sarcopenia research through spatial transcriptomics.Osteoporos Sarcopenia. 2025 Jun;11(2 Suppl):22-31. doi: 10.1016/j.afos.2025.05.002. Epub 2025 Jun 12. Osteoporos Sarcopenia. 2025. PMID: 40718353 Free PMC article. Review.
-
The Impairment of Endothelial Autophagy Accelerates Renal Senescence by Ferroptosis and NLRP3 Inflammasome Signaling Pathways with the Disruption of Endothelial Barrier.Antioxidants (Basel). 2024 Jul 23;13(8):886. doi: 10.3390/antiox13080886. Antioxidants (Basel). 2024. PMID: 39199133 Free PMC article.
-
Dysfunctional β-cell autophagy induces β-cell stress and enhances islet immunogenicity.Front Immunol. 2025 Jan 29;16:1504583. doi: 10.3389/fimmu.2025.1504583. eCollection 2025. Front Immunol. 2025. PMID: 39944686 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

