Thioredoxin is a metabolic rheostat controlling regulatory B cells
- PMID: 38553615
- PMCID: PMC11065695
- DOI: 10.1038/s41590-024-01798-w
Thioredoxin is a metabolic rheostat controlling regulatory B cells
Abstract
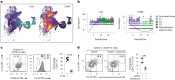
Metabolic programming is important for B cell fate, but the bioenergetic requirement for regulatory B (Breg) cell differentiation and function is unknown. Here we show that Breg cell differentiation, unlike non-Breg cells, relies on mitochondrial electron transport and homeostatic levels of reactive oxygen species (ROS). Single-cell RNA sequencing analysis revealed that TXN, encoding the metabolic redox protein thioredoxin (Trx), is highly expressed by Breg cells, unlike Trx inhibitor TXNIP which was downregulated. Pharmacological inhibition or gene silencing of TXN resulted in mitochondrial membrane depolarization and increased ROS levels, selectively suppressing Breg cell differentiation and function while favoring pro-inflammatory B cell differentiation. Patients with systemic lupus erythematosus (SLE), characterized by Breg cell deficiencies, present with B cell mitochondrial membrane depolarization, elevated ROS and fewer Trx+ B cells. Exogenous Trx stimulation restored Breg cells and mitochondrial membrane polarization in SLE B cells to healthy B cell levels, indicating Trx insufficiency underlies Breg cell impairment in patients with SLE.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
- BB/P504725/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/T002212/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- MRF-057-0004-RG-MCDO-C0800/RCUK | MRC | Medical Research Foundation
- MR/L01257X/2/RCUK | Medical Research Council (MRC)
- FS/SBSRF/22/31031/British Heart Foundation (BHF)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

