Generation of glioblastoma in mice engrafted with human cytomegalovirus-infected astrocytes
- PMID: 38553638
- PMCID: PMC11257955
- DOI: 10.1038/s41417-024-00767-7
Generation of glioblastoma in mice engrafted with human cytomegalovirus-infected astrocytes
Abstract
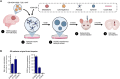
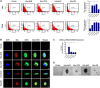
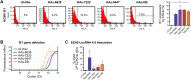
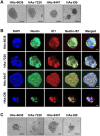
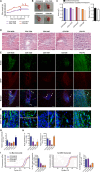
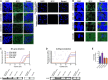
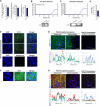
Mounting evidence is identifying human cytomegalovirus (HCMV) as a potential oncogenic virus. HCMV has been detected in glioblastoma multiforme (GB). Herewith, we present the first experimental evidence for the generation of CMV-Elicited Glioblastoma Cells (CEGBCs) possessing glioblastoma-like traits that lead to the formation of glioblastoma in orthotopically xenografted mice. In addition to the already reported oncogenic HCMV-DB strain, we isolated three HCMV clinical strains from GB tissues that transformed HAs toward CEGBCs and generated spheroids from CEGBCs that resulted in the appearance of glioblastoma-like tumors in xenografted mice. These tumors were nestin-positive mostly in the invasive part surrounded by GFAP-positive reactive astrocytes. The glioblastoma immunohistochemistry phenotype was confirmed by EGFR and cMet gene amplification in the tumor parallel to the detection of HCMV IE and UL69 genes and proteins. Our results fit with an HCMV-induced glioblastoma model of oncogenesis in vivo which will open the door to new therapeutic approaches and assess the anti-HCMV treatment as well as immunotherapy in fighting GB which is characterized by poor prognosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Geder L, Sanford EJ, Rohner TJ, Rapp F. Cytomegalovirus and cancer of the prostate: in vitro transformation of human cells. Cancer Treat Rep. 1977;61:139–46. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

