Towards a unified medical microbiome ecology of the OMU for metagenomes and the OTU for microbes
- PMID: 38553666
- PMCID: PMC10979563
- DOI: 10.1186/s12859-023-05591-8
Towards a unified medical microbiome ecology of the OMU for metagenomes and the OTU for microbes
Abstract
Background: Metagenomic sequencing technologies offered unprecedented opportunities and also challenges to microbiology and microbial ecology particularly. The technology has revolutionized the studies of microbes and enabled the high-profile human microbiome and earth microbiome projects. The terminology-change from microbes to microbiomes signals that our capability to count and classify microbes (microbiomes) has achieved the same or similar level as we can for the biomes (macrobiomes) of plants and animals (macrobes). While the traditional investigations of macrobiomes have usually been conducted through naturalists' (Linnaeus & Darwin) naked eyes, and aerial and satellite images (remote-sensing), the large-scale investigations of microbiomes have been made possible by DNA-sequencing-based metagenomic technologies. Two major types of metagenomic sequencing technologies-amplicon sequencing and whole-genome (shotgun sequencing)-respectively generate two contrastingly different categories of metagenomic reads (data)-OTU (operational taxonomic unit) tables representing microorganisms and OMU (operational metagenomic unit), a new term coined in this article to represent various cluster units of metagenomic genes.
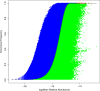
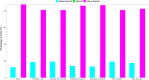
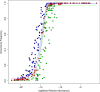
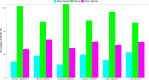
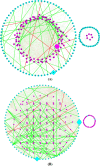
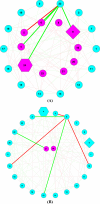
Results: The ecological science of microbiomes based on the OTU representing microbes has been unified with the classic ecology of macrobes (macrobiomes), but the unification based on OMU representing metagenomes has been rather limited. In a previous series of studies, we have demonstrated the applications of several classic ecological theories (diversity, composition, heterogeneity, and biogeography) to the studies of metagenomes. Here I push the envelope for the unification of OTU and OMU again by demonstrating the applications of metacommunity assembly and ecological networks to the metagenomes of human gut microbiomes. Specifically, the neutral theory of biodiversity (Sloan's near neutral model), Ning et al.stochasticity framework, core-periphery network, high-salience skeleton network, special trio-motif, and positive-to-negative ratio are applied to analyze the OMU tables from whole-genome sequencing technologies, and demonstrated with seven human gut metagenome datasets from the human microbiome project.
Conclusions: All of the ecological theories demonstrated previously and in this article, including diversity, composition, heterogeneity, stochasticity, and complex network analyses, are equally applicable to OMU metagenomic analyses, just as to OTU analyses. Consequently, I strongly advocate the unification of OTU/OMU (microbiomes) with classic ecology of plants and animals (macrobiomes) in the context of medical ecology.
Keywords: Core/periphery network; High-salience skeleton network; Medical ecology; Metagenome ecology; Operational metagenomic unit (OMU); Operational taxonomic unit (OTU); Sloan near neutral model; Unified ecology of metagenomes and organisms (species); Unified ecology of microbiomes and macrobiomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Assessing and Interpreting the Metagenome Heterogeneity With Power Law.Front Microbiol. 2020 May 6;11:648. doi: 10.3389/fmicb.2020.00648. eCollection 2020. Front Microbiol. 2020. PMID: 32435232 Free PMC article.
-
Metagenome comparison (MC): A new framework for detecting unique/enriched OMUs (operational metagenomic units) derived from whole-genome sequencing reads.Comput Biol Med. 2024 Sep;180:108852. doi: 10.1016/j.compbiomed.2024.108852. Epub 2024 Aug 12. Comput Biol Med. 2024. PMID: 39137667
-
Measuring metagenome diversity and similarity with Hill numbers.Mol Ecol Resour. 2018 Nov;18(6):1339-1355. doi: 10.1111/1755-0998.12923. Epub 2018 Jul 27. Mol Ecol Resour. 2018. PMID: 29985552
-
Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review.
-
Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses.Microb Genom. 2020 Aug;6(8):mgen000409. doi: 10.1099/mgen.0.000409. Epub 2020 Jul 24. Microb Genom. 2020. PMID: 32706331 Free PMC article. Review.
References
-
- Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton: Princeton University Press; 2001. - PubMed
-
- Duret L. Neutral theory: the null hypothesis of molecular evolution. Nature Education. 2008;1(1):218.
-
- Hartl DL, Clark AG. Principles of population genetics. 3. Sunderland: Sinauer Associates; 1997.
MeSH terms
LinkOut - more resources
Full Text Sources

