FixNCut: single-cell genomics through reversible tissue fixation and dissociation
- PMID: 38553769
- PMCID: PMC10979608
- DOI: 10.1186/s13059-024-03219-5
FixNCut: single-cell genomics through reversible tissue fixation and dissociation
Abstract
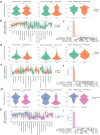
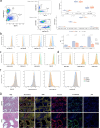
The use of single-cell technologies for clinical applications requires disconnecting sampling from downstream processing steps. Early sample preservation can further increase robustness and reproducibility by avoiding artifacts introduced during specimen handling. We present FixNCut, a methodology for the reversible fixation of tissue followed by dissociation that overcomes current limitations. We applied FixNCut to human and mouse tissues to demonstrate the preservation of RNA integrity, sequencing library complexity, and cellular composition, while diminishing stress-related artifacts. Besides single-cell RNA sequencing, FixNCut is compatible with multiple single-cell and spatial technologies, making it a versatile tool for robust and flexible study designs.
Keywords: Cellular stress; RNA sequencing; Sample fixation; Single-cell genomics; Tissue dissociation.
© 2024. Crown.
Conflict of interest statement
H.H. is a co-founder and shareholder of Omniscope, a scientific advisory board member of MiRXES and Nanostring, and a consultant to Moderna and Singularity. L.G.M is an advisor and shareholder of Omniscope and advisor for ArgenTAG, Millenium Sciences, and Truckee Applied Genomics. J.C.N. is a consultant of Omniscope. M.C.N. has received grants (unrestricted) from GSK, the European Commission, and Lung Foundation Netherlands. M.v.d.B. has received grants (unrestricted) from AstraZeneca, Novartis, GlaxoSmithKline, Roche, Genentech, and Sanofi. F.T. received speaker’s fees from Falk, Janssen, and AbbVie. S.S. has been a consultant for AbbVie, Bristol Myers Squibb, Boehringer Ingelheim, Ferring, Genentech/Roche, Janssen, Lilly, Novartis, Merck Sharp Dohme, Medimmune/AstraZeneca, Pfizer, Protagonist, Sanofi, Takeda, Theravance, and UCB and is a paid speaker for AbbVie, Ferring, Janssen, Merck Sharp Dohme, Novartis, Takeda, and UCB. P.R. has been a consultant for Takeda and Omass. I.A.P. holds current research grants with AstraZeneca and BMS. Omniscope has filed a patent related to the application of the FixNCut protocol. All other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

