Critical steps in the assembly process of the bacterial 50S ribosomal subunit
- PMID: 38554105
- PMCID: PMC11077053
- DOI: 10.1093/nar/gkae199
Critical steps in the assembly process of the bacterial 50S ribosomal subunit
Abstract
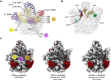
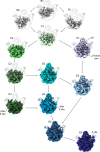
During assembly, ribosomal particles in bacteria fold according to energy landscapes comprised of multiple parallel pathways. Cryo-electron microscopy studies have identified a critical maturation step that occurs during the late assembly stages of the 50S subunit in Bacillus subtilis. This step acts as a point of convergency for all the parallel assembly pathways of the subunit, where an assembly intermediate accumulates in a 'locked' state, causing maturation to pause. Assembly factors then act on this critical step to 'unlock' the last maturation steps involving the functional sites. Without these factors, the 50S subunit fails to complete its assembly, causing cells to die due to a lack of functional ribosomes to synthesize proteins. In this review, we analyze these findings in B. subtilis and examine other cryo-EM studies that have visualized assembly intermediates in different bacterial species, to determine if convergency points in the ribosome assembly process are a common theme among bacteria. There are still gaps in our knowledge, as these methodologies have not yet been applied to diverse species. However, identifying and characterizing these convergency points can reveal how different bacterial species implement unique mechanisms to regulate critical steps in the ribosome assembly process.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






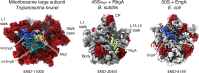
Similar articles
-
RbgA ensures the correct timing in the maturation of the 50S subunits functional sites.Nucleic Acids Res. 2022 Oct 28;50(19):10801-10816. doi: 10.1093/nar/gkac059. Nucleic Acids Res. 2022. PMID: 35141754 Free PMC article.
-
The binding of RbgA to a critical 50S assembly intermediate facilitates YphC function in bacterial ribosomal assembly.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1197. doi: 10.1093/nar/gkae1197. Nucleic Acids Res. 2025. PMID: 39658043 Free PMC article.
-
YphC and YsxC GTPases assist the maturation of the central protuberance, GTPase associated region and functional core of the 50S ribosomal subunit.Nucleic Acids Res. 2016 Sep 30;44(17):8442-55. doi: 10.1093/nar/gkw678. Epub 2016 Aug 2. Nucleic Acids Res. 2016. PMID: 27484475 Free PMC article.
-
Towards an elucidation of the roles of the ribosome during different growth phases in Bacillus subtilis.Biosci Biotechnol Biochem. 2010;74(3):451-61. doi: 10.1271/bbb.90859. Epub 2010 Mar 7. Biosci Biotechnol Biochem. 2010. PMID: 20208344 Review.
-
An emerging mechanism for the maturation of the Small Subunit Processome.Curr Opin Struct Biol. 2022 Apr;73:102331. doi: 10.1016/j.sbi.2022.102331. Epub 2022 Feb 14. Curr Opin Struct Biol. 2022. PMID: 35176592 Review.
Cited by
-
A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.Nucleic Acids Res. 2024 Aug 12;52(14):8483-8499. doi: 10.1093/nar/gkae399. Nucleic Acids Res. 2024. PMID: 38811035 Free PMC article.
-
Assembly of the Bacterial Ribosome with Circularly Permuted rRNA.bioRxiv [Preprint]. 2024 Apr 10:2024.04.10.588894. doi: 10.1101/2024.04.10.588894. bioRxiv. 2024. Update in: Nucleic Acids Res. 2024 Oct 14;52(18):11254-11265. doi: 10.1093/nar/gkae636. PMID: 38644992 Free PMC article. Updated. Preprint.
-
Translational impacts of enzymes that modify ribosomal RNA around the peptidyl transferase centre.RNA Biol. 2024 Jan;21(1):31-41. doi: 10.1080/15476286.2024.2368305. Epub 2024 Jul 1. RNA Biol. 2024. PMID: 38952121 Free PMC article.
-
Assembly of the bacterial ribosome with circularly permuted rRNA.Nucleic Acids Res. 2024 Oct 14;52(18):11254-11265. doi: 10.1093/nar/gkae636. Nucleic Acids Res. 2024. PMID: 39036963 Free PMC article.
-
The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.Nat Commun. 2025 Jan 10;16(1):567. doi: 10.1038/s41467-025-55836-8. Nat Commun. 2025. PMID: 39794318 Free PMC article.
References
-
- Traub P., Nomura M.. Studies on the assembly of ribosomes in vitro. Cold Spring Harb. Symp. Quant. Biol. 1969; 34:63–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

