The CHK1 inhibitor prexasertib in BRCA wild-type platinum-resistant recurrent high-grade serous ovarian carcinoma: a phase 2 trial
- PMID: 38555285
- PMCID: PMC10981752
- DOI: 10.1038/s41467-024-47215-6
The CHK1 inhibitor prexasertib in BRCA wild-type platinum-resistant recurrent high-grade serous ovarian carcinoma: a phase 2 trial
Abstract
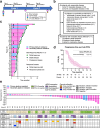
The multi-cohort phase 2 trial NCT02203513 was designed to evaluate the clinical activity of the CHK1 inhibitor (CHK1i) prexasertib in patients with breast or ovarian cancer. Here we report the activity of CHK1i in platinum-resistant high-grade serous ovarian carcinoma (HGSOC) with measurable and biopsiable disease (cohort 5), or without biopsiable disease (cohort 6). The primary endpoint was objective response rate (ORR). Secondary outcomes were safety and progression-free survival (PFS). 49 heavily pretreated patients were enrolled (24 in cohort 5, 25 in cohort 6). Among the 39 RECISTv1.1-evaluable patients, ORR was 33.3% in cohort 5 and 28.6% in cohort 6. Primary endpoint was not evaluable due to early stop of the trial. The median PFS was 4 months in cohort 5 and 6 months in cohort 6. Toxicity was manageable. Translational research was an exploratory endpoint. Potential biomarkers were investigated using pre-treatment fresh biopsies and serial blood samples. Transcriptomic analysis revealed high levels of DNA replication-related genes (POLA1, POLE, GINS3) associated with lack of clinical benefit [defined post-hoc as PFS < 6 months]. Subsequent preclinical experiments demonstrated significant cytotoxicity of POLA1 silencing in combination with CHK1i in platinum-resistant HGSOC cell line models. Therefore, POLA1 expression may be predictive for CHK1i resistance, and the concurrent POLA1 inhibition may improve the efficacy of CHK1i monotherapy in this hard-to-treat population, deserving further investigation.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
J.-M.L. has research grant funding from AstraZeneca and Acrivon Therapeutics (paid to institution) and is on the Scientific Advisory Board of Acrivon Therapeutics and Genentech (unpaid). E.M.S. is on the Data and Safety Monitoring Board of Novartis and the Scientific Advisory Board of Ideaya Bioscience. The other authors have no competing interests to declare.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

