Whole transcriptome sequencing analyses of islets reveal ncRNA regulatory networks underlying impaired insulin secretion and increased β-cell mass in high fat diet-induced diabetes mellitus
- PMID: 38557554
- PMCID: PMC10984535
- DOI: 10.1371/journal.pone.0300965
Whole transcriptome sequencing analyses of islets reveal ncRNA regulatory networks underlying impaired insulin secretion and increased β-cell mass in high fat diet-induced diabetes mellitus
Abstract
Aim: Our study aims to identify novel non-coding RNA-mRNA regulatory networks associated with β-cell dysfunction and compensatory responses in obesity-related diabetes.
Methods: Glucose metabolism, islet architecture and secretion, and insulin sensitivity were characterized in C57BL/6J mice fed on a 60% high-fat diet (HFD) or control for 24 weeks. Islets were isolated for whole transcriptome sequencing to identify differentially expressed (DE) mRNAs, miRNAs, IncRNAs, and circRNAs. Regulatory networks involving miRNA-mRNA, lncRNA-mRNA, and lncRNA-miRNA-mRNA were constructed and functions were assessed through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
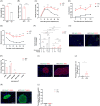
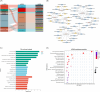
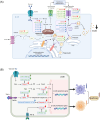
Results: Despite compensatory hyperinsulinemia and a significant increase in β-cell mass with a slow rate of proliferation, HFD mice exhibited impaired glucose tolerance. In isolated islets, insulin secretion in response to glucose and palmitic acid deteriorated after 24 weeks of HFD. Whole transcriptomic sequencing identified a total of 1324 DE mRNAs, 14 DE miRNAs, 179 DE lncRNAs, and 680 DE circRNAs. Our transcriptomic dataset unveiled several core regulatory axes involved in the impaired insulin secretion in HFD mice, such as miR-6948-5p/Cacna1c, miR-6964-3p/Cacna1b, miR-3572-5p/Hk2, miR-3572-5p/Cckar and miR-677-5p/Camk2d. Additionally, proliferative and apoptotic targets, including miR-216a-3p/FKBP5, miR-670-3p/Foxo3, miR-677-5p/RIPK1, miR-802-3p/Smad2 and ENSMUST00000176781/Caspase9 possibly contribute to the increased β-cell mass in HFD islets. Furthermore, competing endogenous RNAs (ceRNA) regulatory network involving 7 DE miRNAs, 15 DE lncRNAs and 38 DE mRNAs might also participate in the development of HFD-induced diabetes.
Conclusions: The comprehensive whole transcriptomic sequencing revealed novel non-coding RNA-mRNA regulatory networks associated with impaired insulin secretion and increased β-cell mass in obesity-related diabetes.
Copyright: © 2024 Ma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Identification of potential biomarkers and pathways associated with carotid atherosclerotic plaques in type 2 diabetes mellitus: A transcriptomics study.Front Endocrinol (Lausanne). 2022 Sep 16;13:981100. doi: 10.3389/fendo.2022.981100. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36187128 Free PMC article.
-
Whole-Transcriptome RNA Sequencing Uncovers the Global Expression Changes and RNA Regulatory Networks in Duck Embryonic Myogenesis.Int J Mol Sci. 2023 Nov 16;24(22):16387. doi: 10.3390/ijms242216387. Int J Mol Sci. 2023. PMID: 38003577 Free PMC article.
-
Whole transcriptome analysis of HCT-8 cells infected by Cryptosporidium parvum.Parasit Vectors. 2022 Nov 24;15(1):441. doi: 10.1186/s13071-022-05565-4. Parasit Vectors. 2022. PMID: 36434735 Free PMC article.
-
Integrative analysis of miRNA-mRNA expression in the brain during high temperature-induced masculinization of female Nile tilapia (Oreochromis niloticus).Genomics. 2024 May;116(3):110856. doi: 10.1016/j.ygeno.2024.110856. Epub 2024 May 10. Genomics. 2024. PMID: 38734154 Review.
-
Current and future perspectives on the regulation and functions of miR-545 in cancer development.Cancer Pathog Ther. 2023 Sep 5;2(3):142-154. doi: 10.1016/j.cpt.2023.09.001. eCollection 2024 Jul. Cancer Pathog Ther. 2023. PMID: 39027151 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

