Variants in autophagy genes MTMR12 and FAM134A are putative modifiers of the hepatic phenotype in α1-antitrypsin deficiency
- PMID: 38557779
- PMCID: PMC11407773
- DOI: 10.1097/HEP.0000000000000865
Variants in autophagy genes MTMR12 and FAM134A are putative modifiers of the hepatic phenotype in α1-antitrypsin deficiency
Abstract
Background and aims: In the classical form of α1-antitrypsin deficiency, a misfolded variant α1-antitrypsin Z accumulates in the endoplasmic reticulum of liver cells and causes liver cell injury by gain-of-function proteotoxicity in a sub-group of affected homozygotes but relatively little is known about putative modifiers. Here, we carried out genomic sequencing in a uniquely affected family with an index case of liver failure and 2 homozygous siblings with minimal or no liver disease. Their sequences were compared to sequences in well-characterized cohorts of homozygotes with or without liver disease, and then candidate sequence variants were tested for changes in the kinetics of α1-antitrypsin variant Z degradation in iPS-derived hepatocyte-like cells derived from the affected siblings themselves.
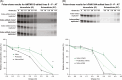
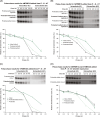
Approach and results: Specific variants in autophagy genes MTMR12 and FAM134A could each accelerate the degradation of α1-antitrypsin variant Z in cells from the index patient, but both MTMR12 and FAM134A variants were needed to slow the degradation of α1-antitrypsin variant Z in cells from a protected sib, indicating that inheritance of both variants is needed to mediate the pathogenic effects of hepatic proteotoxicity at the cellular level. Analysis of homozygote cohorts showed that multiple patient-specific variants in proteostasis genes are likely to explain liver disease susceptibility at the population level.
Conclusions: These results validate the concept that genetic variation in autophagy function can determine susceptibility to liver disease in α1-antitrypsin deficiency and provide evidence that polygenic mechanisms and multiple patient-specific variants are likely needed for proteotoxic pathology.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
Alex Soto-Gutierrez owns stock in Pittsburgh ReLiver and Von Baer Wolff. Adam E. Locke is employed by and owns stock in Regeneron. Michael H. Cho received grants from Bayer. Edwin K Silverman received grants from Bayer and Northpond Laboratories. Ira J. Fox consults for Miromatrix. He owns stock in Pittsburgh ReLiver and Von Baer Wolff. The remaining authors have no conflicts to report.
Figures




References
-
- Strnad P, McElvaney NG, Lomas DA. Alpha1-antitrypsin deficiency. N Engl J Med. 2020;382:1443–1455. - PubMed
-
- Chu AS, Chopra KB, Perlmutter DH. Is severe progressive liver disease caused by alpha-1-antitrypsin deficiency more common in children or adults? Liver Transpl. 2016;22:886–894. - PubMed
-
- Sveger T. Liver disease in alpha1-antitrypsin deficiency detected by screening of 200,000 infants. N Engl J Med. 1976;294:1316–1321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

