This is a preprint.
A Foxf1-Wnt-Nr2f1 cascade promotes atrial cardiomyocyte differentiation in zebrafish
- PMID: 38558972
- PMCID: PMC10980076
- DOI: 10.1101/2024.03.13.584759
A Foxf1-Wnt-Nr2f1 cascade promotes atrial cardiomyocyte differentiation in zebrafish
Update in
-
A Foxf1-Wnt-Nr2f1 cascade promotes atrial cardiomyocyte differentiation in zebrafish.PLoS Genet. 2024 Nov 4;20(11):e1011222. doi: 10.1371/journal.pgen.1011222. eCollection 2024 Nov. PLoS Genet. 2024. PMID: 39495809 Free PMC article.
Abstract
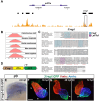
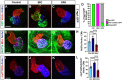
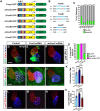
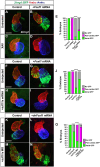
Nr2f transcription factors (TFs) are conserved regulators of vertebrate atrial cardiomyocyte (AC) differentiation. However, little is known about the mechanisms directing Nr2f expression in ACs. Here, we identified a conserved enhancer 3' to the nr2f1a locus, which we call 3'reg1-nr2f1a (3'reg1), that can promote Nr2f1a expression in ACs. Sequence analysis of the enhancer identified putative Lef/Tcf and Foxf TF binding sites. Mutation of the Lef/Tcf sites within the 3'reg1 reporter, knockdown of Tcf7l1a, and manipulation of canonical Wnt signaling support that Tcf7l1a is derepressed via Wnt signaling to activate the transgenic enhancer and promote AC differentiation. Similarly, mutation of the Foxf binding sites in the 3'reg1 reporter, coupled with gain- and loss-of-function analysis supported that Foxf1 promotes expression of the enhancer and AC differentiation. Functionally, we find that Wnt signaling acts downstream of Foxf1 to promote expression of the 3'reg1 reporter within ACs and, importantly, both Foxf1 and Wnt signaling require Nr2f1a to promote a surplus of differentiated ACs. CRISPR-mediated deletion of the endogenous 3'reg1 abrogates the ability of Foxf1 and Wnt signaling to produce surplus ACs in zebrafish embryos. Together, our data support that downstream members of a conserved regulatory network involving Wnt signaling and Foxf1 function on a nr2f1a enhancer to promote AC differentiation in the zebrafish heart.
Figures



References
-
- Bruneau BG, Riley P. Heart Development and Disease. Cold Spring Harbor Laboratory Press; 2020. Available: https://play.google.com/store/books/details?id=pwE6yAEACAAJ
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
