This is a preprint.
Long-read single-cell RNA sequencing enables the study of cancer subclone-specific genotype and phenotype in chronic lymphocytic leukemia
- PMID: 38559060
- PMCID: PMC10979946
- DOI: 10.1101/2024.03.15.585298
Long-read single-cell RNA sequencing enables the study of cancer subclone-specific genotype and phenotype in chronic lymphocytic leukemia
Update in
-
Long-read single-cell RNA sequencing enables the study of cancer subclone-specific genotypes and phenotypes in chronic lymphocytic leukemia.Genome Res. 2025 Apr 14;35(4):686-697. doi: 10.1101/gr.279049.124. Genome Res. 2025. PMID: 39965935 Free PMC article.
Abstract
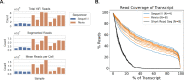
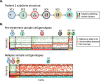
Bruton's tyrosine kinase (BTK) inhibitors are effective for the treatment of chronic lymphocytic leukemia (CLL) due to BTK's role in B cell survival and proliferation. Treatment resistance is most commonly caused by the emergence of the hallmark BTKC481S mutation that inhibits drug binding. In this study, we aimed to investigate whether the presence of additional CLL driver mutations in cancer subclones harboring a BTKC481S mutation accelerates subclone expansion. In addition, we sought to determine whether BTK-mutated subclones exhibit distinct transcriptomic behavior when compared to other cancer subclones. To achieve these goals, we employ our recently published method (Qiao et al. 2024) that combines bulk DNA sequencing and single-cell RNA sequencing (scRNA-seq) data to genotype individual cells for the presence or absence of subclone-defining mutations. While the most common approach for scRNA-seq includes short-read sequencing, transcript coverage is limited due to the vast majority of the reads being concentrated at the priming end of the transcript. Here, we utilized MAS-seq, a long-read scRNAseq technology, to substantially increase transcript coverage across the entire length of the transcripts and expand the set of informative mutations to link cells to cancer subclones in six CLL patients who acquired BTKC481S mutations during BTK inhibitor treatment. We found that BTK-mutated subclones often acquire additional mutations in CLL driver genes, leading to faster subclone proliferation. When examining subclone-specific gene expression, we found that in one patient, BTK-mutated subclones are transcriptionally distinct from the rest of the malignant B cell population with an overexpression of CLL-relevant genes.
Conflict of interest statement
Competing interest statement D.M.S has received research funding from Abbvie, AstraZeneca, Genentech, and Novartis and is a consultant for Abbvie, AstraZeneca, Beigene, Celgene, Eli Lilly, Genentech, Janssen, Pharmacyclic. J.A.W. has received research funding from Abbvie, Janssen, Pharmacyclics, and Schrodinger and is a consultant for Abbvie, AstraZeneca, Beigene, Genentech, Janssen, Loxo/Lilly, Merck, Newave, Pharmacyclics. The remaining authors declare no competing financial interests.
Figures

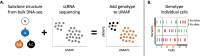


References
-
- Arruga F, Gizdic B, Serra S, Vaisitti T, Ciardullo C, Coscia M, Laurenti L, D’Arena G, Jaksic O, Inghirami G, et al. 2014. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia 28: 1060–1070. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
