This is a preprint.
Molecular Mechanisms of Genotype-Dependent Lifespan Variation Mediated by Caloric Restriction: Insight from Wild Yeast Isolates
- PMID: 38559208
- PMCID: PMC10979966
- DOI: 10.1101/2024.03.17.585422
Molecular Mechanisms of Genotype-Dependent Lifespan Variation Mediated by Caloric Restriction: Insight from Wild Yeast Isolates
Update in
-
Molecular mechanisms of genotype-dependent lifespan variation mediated by caloric restriction: insight from wild yeast isolates.Front Aging. 2024 Jul 11;5:1408160. doi: 10.3389/fragi.2024.1408160. eCollection 2024. Front Aging. 2024. PMID: 39055969 Free PMC article.
Abstract
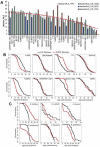
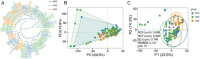
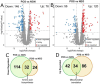
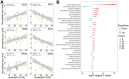
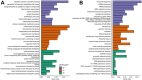
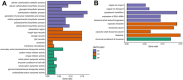
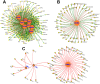
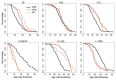
Caloric restriction (CR) is known to extend lifespan across different species and holds great promise for preventing human age-onset pathologies. However, two major challenges exist. First, despite extensive research, the mechanisms of lifespan extension in response to CR remain elusive. Second, genetic differences causing variations in response to CR and genetic factors contributing to variability of CR response on lifespan are largely unknown. Here, we took advantage of natural genetic variation across 46 diploid wild yeast isolates of Saccharomyces species and the lifespan variation under CR conditions to uncover the molecular factors associated with CR response types. We identified genes and metabolic pathways differentially regulated in CR-responsive versus non-responsive strains. Our analysis revealed that altered mitochondrial function and activation of GCN4-mediated environmental stress response are inevitably linked to lifespan variation in response to CR and a unique mitochondrial metabolite might be utilized as a predictive marker for CR response rate. In sum, our data suggests that the effects of CR on longevity may not be universal, even among the closely related species or strains of a single species. Since mitochondrial-mediated signaling pathways are evolutionarily conserved, the dissection of related genetic pathways will be relevant to understanding the mechanism by which CR elicits its longevity effect.
Keywords: Caloric restriction; genotype variation; lifespan; metabolism; yeast.
Conflict of interest statement
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Molecular mechanisms of genotype-dependent lifespan variation mediated by caloric restriction: insight from wild yeast isolates.Front Aging. 2024 Jul 11;5:1408160. doi: 10.3389/fragi.2024.1408160. eCollection 2024. Front Aging. 2024. PMID: 39055969 Free PMC article.
-
Characterization of global gene expression during assurance of lifespan extension by caloric restriction in budding yeast.Exp Gerontol. 2013 Dec;48(12):1455-68. doi: 10.1016/j.exger.2013.10.001. Epub 2013 Oct 11. Exp Gerontol. 2013. PMID: 24126084
-
Patterns of intraspecific variability in the response to caloric restriction.Exp Gerontol. 2014 Mar;51:28-37. doi: 10.1016/j.exger.2013.12.005. Epub 2013 Dec 31. Exp Gerontol. 2014. PMID: 24384399 Free PMC article.
-
The fine line between lifespan extension and shortening in response to caloric restriction.Nucleus. 2014 Jan-Feb;5(1):56-65. doi: 10.4161/nucl.27929. Epub 2014 Jan 27. Nucleus. 2014. PMID: 24637399 Free PMC article. Review.
-
Caloric restriction and the aging process: a critique.Free Radic Biol Med. 2014 Aug;73:366-82. doi: 10.1016/j.freeradbiomed.2014.05.015. Epub 2014 Jun 2. Free Radic Biol Med. 2014. PMID: 24941891 Free PMC article. Review.
References
-
- Sinclair D. A. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev 126, 987–1002 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
