This is a preprint.
Autophagosome turnover requires Arp2/3 complex-mediated maintenance of lysosomal integrity
- PMID: 38559247
- PMCID: PMC10980047
- DOI: 10.1101/2024.03.12.584718
Autophagosome turnover requires Arp2/3 complex-mediated maintenance of lysosomal integrity
Abstract
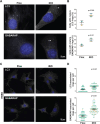
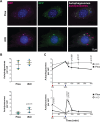
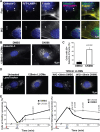
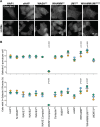
Autophagy is an intracellular degradation process that maintains homeostasis, responds to stress, and plays key roles in the prevention of aging and disease. Autophagosome biogenesis, vesicle rocketing, and autolysosome tubulation are controlled by multiple actin nucleation factors, but the impact of actin assembly on completion of the autophagic pathway is not well understood. Here we studied autophagosome and lysosome remodeling in fibroblasts harboring an inducible knockout (iKO) of the Arp2/3 complex, an essential actin nucleator. Arp2/3 complex ablation resulted in increased basal levels of autophagy receptors and lipidated membrane proteins from the LC3 and GABARAP families. Under both steady-state and starvation conditions, Arp2/3 iKO cells accumulated abnormally high numbers of autolysosomes, suggesting a defect in autophagic flux. The inability of Arp2/3 complex-deficient cells to complete autolysosome degradation and turnover is explained by the presence of damaged, leaky lysosomes. In cells treated with an acute lysosomal membrane-damaging agent, the Arp2/3-activating protein WHAMM is recruited to lysosomes, where Arp2/3 complex-dependent actin assembly is crucial for restoring intact lysosomal structure. These results establish the Arp2/3 complex as a central player late in the canonical autophagy pathway and reveal a new role for the actin nucleation machinery in maintaining lysosomal integrity.
Figures





Similar articles
-
WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism.Curr Biol. 2015 Jun 29;25(13):1791-7. doi: 10.1016/j.cub.2015.05.042. Epub 2015 Jun 18. Curr Biol. 2015. PMID: 26096974 Free PMC article.
-
WHAMM links actin assembly via the Arp2/3 complex to autophagy.Autophagy. 2015;11(9):1702-4. doi: 10.1080/15548627.2015.1073434. Autophagy. 2015. PMID: 26291929 Free PMC article.
-
WHAMM functions in kidney reabsorption and polymerizes actin to promote autophagosomal membrane closure and cargo sequestration.bioRxiv [Preprint]. 2024 Jan 23:2024.01.22.576497. doi: 10.1101/2024.01.22.576497. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Jun 1;35(6):ar80. doi: 10.1091/mbc.E24-01-0025. PMID: 38328079 Free PMC article. Updated. Preprint.
-
Regulation of actin nucleation and autophagosome formation.Cell Mol Life Sci. 2016 Sep;73(17):3249-63. doi: 10.1007/s00018-016-2224-z. Epub 2016 May 4. Cell Mol Life Sci. 2016. PMID: 27147468 Free PMC article. Review.
-
NPFs-mediated actin cytoskeleton: a new viewpoint on autophagy regulation.Cell Commun Signal. 2024 Feb 12;22(1):111. doi: 10.1186/s12964-023-01444-2. Cell Commun Signal. 2024. PMID: 38347641 Free PMC article. Review.
References
-
- Aplin A., Jasionowski T., Tuttle D.L., Lenk S.E., and Dunn W.A. Jr. (1992). Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. Journal of Cellular Physiology 152, 458–466. - PubMed
-
- Bhattacharya A., Mukherjee R., Kuncha S.K., Brunstein M.E., Rathore R., Junek S., Münch C., and Dikic I. (2023). A lysosome membrane regeneration pathway depends on TBC1D15 and autophagic lysosomal reformation proteins. Nature Cell Biology 25, 685–698. - PubMed
-
- Boyle K.B., and Randow F. (2013). The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Current Opinion in Microbiology 16, 339–348. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
