This is a preprint.
PPIscreenML: Structure-based screening for protein-protein interactions using AlphaFold
- PMID: 38559274
- PMCID: PMC10979958
- DOI: 10.1101/2024.03.16.585347
PPIscreenML: Structure-based screening for protein-protein interactions using AlphaFold
Abstract
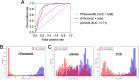
Protein-protein interactions underlie nearly all cellular processes. With the advent of protein structure prediction methods such as AlphaFold2 (AF2), models of specific protein pairs can be built extremely accurately in most cases. However, determining the relevance of a given protein pair remains an open question. It is presently unclear how to use best structure-based tools to infer whether a pair of candidate proteins indeed interact with one another: ideally, one might even use such information to screen amongst candidate pairings to build up protein interaction networks. Whereas methods for evaluating quality of modeled protein complexes have been co-opted for determining which pairings interact (e.g., pDockQ and iPTM), there have been no rigorously benchmarked methods for this task. Here we introduce PPIscreenML, a classification model trained to distinguish AF2 models of interacting protein pairs from AF2 models of compelling decoy pairings. We find that PPIscreenML out-performs methods such as pDockQ and iPTM for this task, and further that PPIscreenML exhibits impressive performance when identifying which ligand/receptor pairings engage one another across the structurally conserved tumor necrosis factor superfamily (TNFSF). Analysis of benchmark results using complexes not seen in PPIscreenML development strongly suggest that the model generalizes beyond training data, making it broadly applicable for identifying new protein complexes based on structural models built with AF2.
Figures



Similar articles
-
Rēs loquunt: What's wrong with AlphaFold's score and how to fix it.bioRxiv [Preprint]. 2025 Feb 14:2025.02.10.637595. doi: 10.1101/2025.02.10.637595. bioRxiv. 2025. PMID: 39990437 Free PMC article. Preprint.
-
Blind Assessment of Monomeric AlphaFold2 Protein Structure Models with Experimental NMR Data.bioRxiv [Preprint]. 2023 Jan 22:2023.01.22.525096. doi: 10.1101/2023.01.22.525096. bioRxiv. 2023. Update in: J Magn Reson. 2023 Jul;352:107481. doi: 10.1016/j.jmr.2023.107481. PMID: 36712039 Free PMC article. Updated. Preprint.
-
Blind assessment of monomeric AlphaFold2 protein structure models with experimental NMR data.J Magn Reson. 2023 Jul;352:107481. doi: 10.1016/j.jmr.2023.107481. Epub 2023 May 20. J Magn Reson. 2023. PMID: 37257257 Free PMC article.
-
AlphaFold2 and its applications in the fields of biology and medicine.Signal Transduct Target Ther. 2023 Mar 14;8(1):115. doi: 10.1038/s41392-023-01381-z. Signal Transduct Target Ther. 2023. PMID: 36918529 Free PMC article. Review.
-
A Perspective on the Prospective Use of AI in Protein Structure Prediction.J Chem Inf Model. 2024 Jan 8;64(1):26-41. doi: 10.1021/acs.jcim.3c01361. Epub 2023 Dec 20. J Chem Inf Model. 2024. PMID: 38124369 Review.
References
-
- Choi SG, Olivet J, Cassonnet P, Vidalain PO, Luck K, Lambourne L, Spirohn K, Lemmens I, Dos Santos M, Demeret C, Jones L, Rangarajan S, Bian W, Coutant EP, Janin YL, van der Werf S, Trepte P, Wanker EE, De Las Rivas J, Tavernier J, Twizere JC, Hao T, Hill DE, Vidal M, Calderwood MA, Jacob Y. Maximizing binary interactome mapping with a minimal number of assays. Nat Commun. 2019; 10:3907. - PMC - PubMed
-
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, Yildirim MA, Simonis N, Heinzmann K, Gebreab F, Sahalie JM, Cevik S, Simon C, de Smet AS, Dann E, Smolyar A, Vinayagam A, Yu H, Szeto D, Borick H, Dricot A, Klitgord N, Murray RR, Lin C, Lalowski M, Timm J, Rau K, Boone C, Braun P, Cusick ME, Roth FP, Hill DE, Tavernier J, Wanker EE, Barabasi AL, Vidal M. An empirical framework for binary interactome mapping. Nat Methods. 2009; 6:83–90. - PMC - PubMed
-
- Elhabashy H, Merino F, Alva V, Kohlbacher O, Lupas AN. Exploring protein-protein interactions at the proteome level. Structure. 2022; 30:462–75. - PubMed
-
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989; 340:245–6. - PubMed
-
- Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012; 12:1576–90. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
