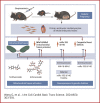
Small Molecule Activators of Mitochondrial Fusion Prevent Congenital Heart Defects Induced by Maternal Diabetes
- PMID: 38559623
- PMCID: PMC10978414
- DOI: 10.1016/j.jacbts.2023.11.008
Small Molecule Activators of Mitochondrial Fusion Prevent Congenital Heart Defects Induced by Maternal Diabetes
Abstract
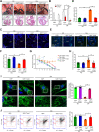
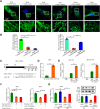
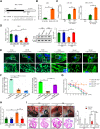
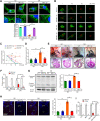
Most congenital heart defect (CHD) cases are attributed to nongenetic factors; however, the mechanisms underlying nongenetic factor-induced CHDs are elusive. Maternal diabetes is one of the nongenetic factors, and this study aimed to determine whether impaired mitochondrial fusion contributes to maternal diabetes-induced CHDs and if mitochondrial fusion activators, teriflunomide and echinacoside, could reduce CHD incidence in diabetic pregnancy. We demonstrated maternal diabetes-activated FoxO3a increases miR-140 and miR-195, which in turn represses Mfn1 and Mfn2, leading to mitochondrial fusion defects and CHDs. Two mitochondrial fusion activators are effective in preventing CHDs in diabetic pregnancy.
Keywords: congenital heart defect; maternal diabetes; microRNA; mitochondrial fusion; mitofusin 1; mitofusin 2.
© 2024 The Authors.
Conflict of interest statement
This work was financially supported by National Institutes of Health grants R01DK083243, R01DK101972, R01DK103024, R01HL131737, R01HL134368, R01HL139060, R01HL153141, and R01HL151108. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures
Similar articles
-
MicroRNA-322 overexpression reduces neural tube defects in diabetic pregnancies.Am J Obstet Gynecol. 2024 Feb;230(2):254.e1-254.e13. doi: 10.1016/j.ajog.2023.07.048. Epub 2023 Jul 31. Am J Obstet Gynecol. 2024. PMID: 37531989 Free PMC article.
-
Mitochondrial Dysfunction and Pharmacodynamics of Mitofusin Activation in Murine Charcot-Marie-Tooth Disease Type 2A.J Pharmacol Exp Ther. 2022 Nov;383(2):137-148. doi: 10.1124/jpet.122.001332. Epub 2022 Sep 2. J Pharmacol Exp Ther. 2022. PMID: 36507849 Free PMC article.
-
Association of certain chronic maternal diseases with the risk of specific congenital heart defects: a population-based study.Eur J Obstet Gynecol Reprod Biol. 2014 Nov;182:1-6. doi: 10.1016/j.ejogrb.2014.08.022. Epub 2014 Aug 27. Eur J Obstet Gynecol Reprod Biol. 2014. PMID: 25216447
-
Say NO to ROS: Their Roles in Embryonic Heart Development and Pathogenesis of Congenital Heart Defects in Maternal Diabetes.Antioxidants (Basel). 2019 Oct 1;8(10):436. doi: 10.3390/antiox8100436. Antioxidants (Basel). 2019. PMID: 31581464 Free PMC article. Review.
-
Risk of congenital heart defects in offspring exposed to maternal diabetes mellitus: an updated systematic review and meta-analysis.Arch Gynecol Obstet. 2019 Dec;300(6):1491-1506. doi: 10.1007/s00404-019-05376-6. Epub 2019 Nov 12. Arch Gynecol Obstet. 2019. PMID: 31713644
Cited by
-
Genetic and Environmental Contributors To Congenital Heart Disease.Curr Treat Options Cardiovasc Med. 2025;27(1):36. doi: 10.1007/s11936-025-01091-5. Epub 2025 May 26. Curr Treat Options Cardiovasc Med. 2025. PMID: 40438121 Free PMC article. Review.
-
Mitochondrial Targeted Therapies to Prevent Maternal Diabetes-Induced Congenital Heart Defects.JACC Basic Transl Sci. 2024 Mar 28;9(3):319-321. doi: 10.1016/j.jacbts.2024.01.016. eCollection 2024 Mar. JACC Basic Transl Sci. 2024. PMID: 38559633 Free PMC article.
-
Cytochrome P450 2E1 aggravates DXR-induced myocardial injury through imbalanced mitochondrial OPA1.Cell Commun Signal. 2025 Apr 30;23(1):208. doi: 10.1186/s12964-025-02197-w. Cell Commun Signal. 2025. PMID: 40307912 Free PMC article.
References
-
- Mathews T.J., Driscoll A.K. Trends in infant mortality in the United States, 2005-2014. NCHS Data Brief. 2017;(279):1–8. - PubMed
-
- Virani S.S., Alonso A., Benjamin E.J., et al. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141:e139–e596. - PubMed
-
- Botto L.D., Mulinare J., Erickson J.D. Occurrence of congenital heart defects in relation to maternal mulitivitamin use. Am J Epidemiol. 2000;151:878–884. - PubMed
-
- Jenkins K.J., Correa A., Feinstein J.A., et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

