Exposure to air pollution is associated with DNA methylation changes in sperm
- PMID: 38559770
- PMCID: PMC10980975
- DOI: 10.1093/eep/dvae003
Exposure to air pollution is associated with DNA methylation changes in sperm
Abstract
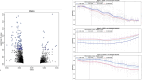
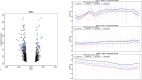
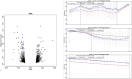
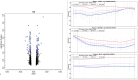
Exposure to air pollutants has been associated with adverse health outcomes in adults and children who were prenatally exposed. In addition to reducing exposure to air pollutants, it is important to identify their biologic targets in order to mitigate the health consequences of exposure. One molecular change associated with prenatal exposure to air pollutants is DNA methylation (DNAm), which has been associated with changes in placenta and cord blood tissues at birth. However, little is known about how air pollution exposure impacts the sperm epigenome, which could provide important insights into the mechanism of transmission to offspring. In the present study, we explored whether exposure to particulate matter less than 2.5 microns in diameter, particulate matter less than 10 microns in diameter, nitrogen dioxide (NO2), or ozone (O3) was associated with DNAm in sperm contributed by participants in the Early Autism Risk Longitudinal Investigation prospective pregnancy cohort. Air pollution exposure measurements were calculated as the average exposure for each pollutant measured within 4 weeks prior to the date of sample collection. Using array-based genome-scale methylation analyses, we identified 80, 96, 35, and 67 differentially methylated regions (DMRs) significantly associated with particulate matter less than 2.5 microns in diameter, particulate matter less than 10 microns in diameter, NO2, and O3, respectively. While no DMRs were associated with exposure to all four pollutants, we found that genes overlapping exposure-related DMRs had a shared enrichment for gene ontology biological processes related to neurodevelopment. Together, these data provide compelling support for the hypothesis that paternal exposure to air pollution impacts DNAm in sperm, particularly in regions implicated in neurodevelopment.
Keywords: DNA methylation; air pollution; epigenetics; genome-scale; sperm.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
Dr Ladd-Acosta reports receiving consulting fees from the University of Iowa for providing expertise on autism spectrum disorder epigenetics outside of this work. No other authors have anything to declare.
Figures
References
-
- Oudin A, Frondelius K, Haglund N et al. Prenatal exposure to air pollution as a potential risk factor for autism and ADHD. Environ Int 2019;133:105149. - PubMed
-
- Breton CV, Yao J, Millstein J et al. Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with newborn LINE1 and alu methylation and childhood blood pressure and carotid intima-media thickness in the children’s health study. Environ Health Perspect 2016;124:1905–12. - PMC - PubMed
-
- Fossati S, Valvi D, Martinez D et al. Prenatal air pollution exposure and growth and cardio-metabolic risk in preschoolers. Environ Int 2020;138:105619. - PubMed
-
- Tingskov Pedersen CE, Eliasen AU, Ketzel M et al. Prenatal exposure to ambient air pollution is associated with early life immune perturbations. J Allergy Clin Immunol 2023;151:212–21. - PubMed

